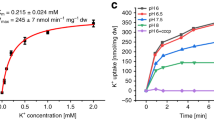
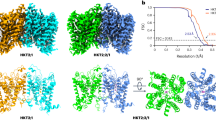
Abstract
In bacteria, archaea, fungi and plants the Trk, Ktr and HKT ion transporters are key components of osmotic regulation, pH homeostasis and resistance to drought and high salinity. These ion transporters are functionally diverse: they can function as Na+ or K+ channels and possibly as cation/K+ symporters. They are closely related to potassium channels both at the level of the membrane protein and at the level of the cytosolic regulatory domains. Here we describe the crystal structure of a Ktr K+ transporter, the KtrAB complex from Bacillus subtilis. The structure shows the dimeric membrane protein KtrB assembled with a cytosolic octameric KtrA ring bound to ATP, an activating ligand. A comparison between the structure of KtrAB–ATP and the structures of the isolated full-length KtrA protein with ATP or ADP reveals a ligand-dependent conformational change in the octameric ring, raising new ideas about the mechanism of activation in these transporters.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Corratgé-Faillie, C. et al. Potassium and sodium transport in non-animal cells: the Trk/Ktr/HKT transporter family. Cell. Mol. Life Sci. 67, 2511–2532 (2010)
Durell, S. R., Hao, Y., Nakamura, T., Bakker, E. P. & Guy, H. R. Evolutionary relationship between K+ channels and symporters. Biophys. J. 77, 775–788 (1999)
Nakamura, T., Yuda, R., Unemoto, T. & Bakker, E. P. KtrAB, a new type of bacterial K+-uptake system from Vibrio alginolyticus. J. Bacteriol. 180, 3491–3494 (1998)
Hänelt, I. et al. KtrB, a member of the superfamily of K+ transporters. Eur. J. Cell Biol. 90, 696–704 (2011)
Tholema, N., Bakker, E. P., Suzuki, A. & Nakamura, T. Change to alanine of one out of four selectivity filter glycines in KtrB causes a two orders of magnitude decrease in the affinities for both K+ and Na+ of the Na+ dependent K+ uptake system KtrAB from Vibrio alginolyticus. FEBS Lett. 450, 217–220 (1999)
Albright, R. A., Joh, K. & Morais-Cabral, J. H. Probing the structure of the dimeric KtrB membrane protein. J. Biol. Chem. 282, 35046–35055 (2007)
Maser, P. et al. Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proc. Natl Acad. Sci. USA 99, 6428–6433 (2002)
Tholema, N. et al. All four putative selectivity filter glycine residues in KtrB are essential for high affinity and selective K+ uptake by the KtrAB system from Vibrio alginolyticus. J. Biol. Chem. 280, 41146–41154 (2005)
Cao, Y. et al. Crystal structure of a potassium ion transporter, TrkH. Nature 471, 336–340 (2011)
Doyle, D. A. et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69–77 (1998)
Jiang, Y. et al. The open pore conformation of potassium channels. Nature 417, 523–526 (2002)
Jiang, Y. et al. Crystal structure and mechanism of a calcium-gated potassium channel. Nature 417, 515–522 (2002)
Wu, Y., Yang, Y., Ye, S. & Jiang, Y. Structure of the gating ring from the human large-conductance Ca2+-gated K+ channel. Nature 466, 393–397 (2010)
Yuan, P., Leonetti, M. D., Pico, A. R., Hsiung, Y. & MacKinnon, R. Structure of the human BK channel Ca2+-activation apparatus at 3.0 Å resolution. Science 329, 182–186 (2010)
Yuan, P., Leonetti, M. D., Hsiung, Y. & MacKinnon, R. Open structure of the Ca2+ gating ring in the high-conductance Ca2+-activated K+ channel. Nature 481, 94–97 (2012)
Holtmann, G., Bakker, E. P., Uozumi, N. & Bremer, E. KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J. Bacteriol. 185, 1289–1298 (2003)
Berry, S. et al. Potassium uptake in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803 mainly depends on a Ktr-like system encoded by slr1509 (ntpJ). FEBS Lett. 548, 53–58 (2003)
Matsuda, N. et al. Na+-dependent K+ uptake Ktr system from the cyanobacterium Synechocystis sp. PCC 6803 and its role in the early phases of cell adaptation to hyperosmotic shock. J. Biol. Chem. 279, 54952–54962 (2004)
Hanelt, I. et al. Gain of function mutations in membrane region M2C2 of KtrB open a gate controlling K+ transport by the KtrAB system from Vibrio alginolyticus. J. Biol. Chem. 285, 10318–10327 (2010)
Kroning, N. et al. ATP binding to the KTN/RCK subunit KtrA from the K+-uptake system KtrAB of Vibrio alginolyticus: its role in the formation of the KtrAB complex and its requirement in vivo. J. Biol. Chem. 282, 14018–14027 (2007)
Albright, R. A., Ibar, J. L., Kim, C. U., Gruner, S. M. & Morais-Cabral, J. H. The RCK domain of the KtrAB K+ transporter: multiple conformations of an octameric ring. Cell 126, 1147–1159 (2006)
Roosild, T. P., Miller, S., Booth, I. R. & Choe, S. A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch. Cell 109, 781–791 (2002)
Heginbotham, L., Kolmakova-Partensky, L. & Miller, C. Functional reconstitution of a prokaryotic K+ channel. J. Gen. Physiol. 111, 741–749 (1998)
Ye, S., Li, Y., Chen, L. & Jiang, Y. Crystal structures of a ligand-free MthK gating ring: insights into the ligand gating mechanism of K+ channels. Cell 126, 1161–1173 (2006)
Olesen, C. et al. The structural basis of calcium transport by the calcium pump. Nature 450, 1036–1042 (2007)
Cuello, L. G. et al. Structural basis for the coupling between activation and inactivation gates in K+ channels. Nature 466, 272–275 (2010)
Trudeau, M. C. Unlocking the mechanisms of HCN channel gating with locked-open and locked-closed channels. J. Gen. Physiol. 140, 457–461 (2012)
Hilf, R. J. & Dutzler, R. A prokaryotic perspective on pentameric ligand-gated ion channel structure. Curr. Opin. Struct. Biol. 19, 418–424 (2009)
Nimigean, C. M. A radioactive uptake assay to measure ion transport across ion channel-containing liposomes. Nature Protocols 1, 1207–1212 (2006)
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010)
Leslie, A. G. The integration of macromolecular diffraction data. Acta Crystallogr. D 62, 48–57 (2006)
Kelley, L. A. & Sternberg, M. J. Protein structure prediction on the Web: a case study using the Phyre server. Nature Protocols 4, 363–371 (2009)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Kleywegt, G. J. & Jones, T. A. Software for handling macromolecular envelopes. Acta Crystallogr. D 55, 941–944 (1999)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 8060–8065 (2006)
Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.3r1 (2010)
Gille, C. & Frommel, C. STRAP: editor for STRuctural Alignments of Proteins. Bioinformatics 17, 377–378 (2001)
Gouet, P., Robert, X. & Courcelle, E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31, 3320–3323 (2003)
Ho, B. K. & Gruswitz, F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49 (2008)
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011)
Acknowledgements
We are grateful for access to ID14-1/ID14-4/ID-29 at ESRF (through the Portuguese BAG), PXII at SLS, XRD1 at ELETTRA and PROXIMA1 at SOLEIL and thank the respective support staff. A.S. was supported by FEBS (Long term fellowship). This work was funded by EMBO (Installation grant), by FEDER funds through the Operational Competitiveness Program–COMPETE and by National Funds through FCT–Fundação para a Ciência e a Tecnologia under the projects FCOMP-01-0124-FEDER-022718 (PEst-C/SAU/LA0002/2011), FCOMP-01-0124-FEDER-009028 (PTDC/BIA-PRO/099861/2008) and FCOMP-01-0124-FEDER-010781 (PTDC/QUI-BIQ/105342/2008). We also thank G. Gabant and M. Cadene at the ‘Plateforme de Spectrometrie de Masse’ at CBM, CNRS, Orleans for mass spectrometry analysis, and C. Harley for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
R.S.V.-P. performed all crystallographic work with the support of J.H.M.-C.; R.S.V.-P. and A.S. performed the functional and biochemical characterization; J.H.M.-C., R.S.V.-P. and A.S. designed experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-10, which illustrates different aspects of the structural and biochemical characterization of the KtrAB potassium transporter, Supplementary Table 1 showing statistics for diffraction data and crystallographic refinement for the KtrAB, KtrA-ATP and KtrA-ADP structures, and a Supplementary Discussion containing detailed information about the contact regions established between the KtrB homodimer and KtrA ring, as well as discussion of the activation models presented in the main text. (PDF 27413 kb)
Rights and permissions
About this article
Cite this article
Vieira-Pires, R., Szollosi, A. & Morais-Cabral, J. The structure of the KtrAB potassium transporter. Nature 496, 323–328 (2013). https://doi.org/10.1038/nature12055
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12055
This article is cited by
-
Dealing with Environmental Fluctuations: Diversity of Potassium Uptake Systems Across the Three Domains of Life
Journal of Plant Growth Regulation (2023)
-
Wie Bakterien die Aufnahme von Kaliumionen regulieren
BIOspektrum (2022)
-
Mechanism of high affinity potassium transporter (HKT) towards improved crop productivity in saline agricultural lands
3 Biotech (2022)
-
Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC
Nature Communications (2021)
-
Barley sodium content is regulated by natural variants of the Na+ transporter HvHKT1;5
Communications Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.