Abstract
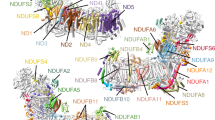
Complex I is the first and largest enzyme of the respiratory chain and has a central role in cellular energy production through the coupling of NADH:ubiquinone electron transfer to proton translocation. It is also implicated in many common human neurodegenerative diseases. Here, we report the first crystal structure of the entire, intact complex I (from Thermus thermophilus) at 3.3 Å resolution. The structure of the 536-kDa complex comprises 16 different subunits, with a total of 64 transmembrane helices and 9 iron–sulphur clusters. The core fold of subunit Nqo8 (ND1 in humans) is, unexpectedly, similar to a half-channel of the antiporter-like subunits. Small subunits nearby form a linked second half-channel, which completes the fourth proton-translocation pathway (present in addition to the channels in three antiporter-like subunits). The quinone-binding site is unusually long, narrow and enclosed. The quinone headgroup binds at the deep end of this chamber, near iron–sulphur cluster N2. Notably, the chamber is linked to the fourth channel by a ‘funnel’ of charged residues. The link continues over the entire membrane domain as a flexible central axis of charged and polar residues, and probably has a leading role in the propagation of conformational changes, aided by coupling elements. The structure suggests that a unique, out-of-the-membrane quinone-reaction chamber enables the redox energy to drive concerted long-range conformational changes in the four antiporter-like domains, resulting in translocation of four protons per cycle.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Walker, J. E. The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q. Rev. Biophys. 25, 253–324 (1992)
Yagi, T. & Matsuno-Yagi, A. The proton-translocating NADH-quinone oxidoreductase in the respiratory chain: the secret unlocked. Biochemistry 42, 2266–2274 (2003)
Brandt, U. Energy converting NADH:quinone oxidoreductase (complex I). Annu. Rev. Biochem. 75, 69–92 (2006)
Ohnishi, T. Iron–sulfur clusters/semiquinones in complex I. Biochim. Biophys. Acta 1364, 186–206 (1998)
Sazanov, L. A. Respiratory complex I: mechanistic and structural insights provided by the crystal structure of the hydrophilic domain. Biochemistry 46, 2275–2288 (2007)
Hirst, J. Towards the molecular mechanism of respiratory complex I. Biochem. J. 425, 327–339 (2010)
Sazanov, L. A. A Structural Perspective on Respiratory Complex I: Structure and Function of NADH:Ubiquinone Oxidoreductase (Springer, 2012)
Galkin, A. S., Grivennikova, V. G. & Vinogradov, A. D. H. H+/2ē stoichiometry in NADH–quinone reductase reactions catalyzed by bovine heart submitochondrial particles. FEBS Lett. 451, 157–161 (1999)
Galkin, A., Drose, S. & Brandt, U. The proton pumping stoichiometry of purified mitochondrial complex I reconstituted into proteoliposomes. Biochim. Biophys. Acta 1757, 1575–1581 (2006)
Moser, C. C., Farid, T. A., Chobot, S. E. & Dutton, P. L. Electron tunneling chains of mitochondria. Biochim. Biophys. Acta 1757, 1096–1109 (2006)
Watt, I. N., Montgomery, M. G., Runswick, M. J., Leslie, A. G. & Walker, J. E. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc. Natl Acad. Sci. USA 107, 16823–16827 (2010)
Vinogradov, A. D. Catalytic properties of the mitochondrial NADH–ubiquinone oxidoreductase (complex I) and the pseudo-reversible active/inactive enzyme transition. Biochim. Biophys. Acta 1364, 169–185 (1998)
Schapira, A. H. Human complex I defects in neurodegenerative diseases. Biochim. Biophys. Acta 1364, 261–270 (1998)
Murphy, M. P. How mitochondria produce reactive oxygen species. Biochem. J. 417, 1–13 (2009)
Dawson, T. M. & Dawson, V. L. Molecular pathways of neurodegeneration in Parkinson’s disease. Science 302, 819–822 (2003)
Balaban, R. S., Nemoto, S. & Finkel, T. Mitochondria, oxidants, and aging. Cell 120, 483–495 (2005)
Carroll, J. et al. Bovine complex I is a complex of 45 different subunits. J. Biol. Chem. 281, 32724–32727 (2006)
Balsa, E. et al. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 16, 378–386 (2012)
Yip, C. Y., Harbour, M. E., Jayawardena, K., Fearnley, I. M. & Sazanov, L. A. Evolution of respiratory complex I: “supernumerary” subunits are present in the α-proteobacterial enzyme. J. Biol. Chem. 286, 5023–5033 (2011)
Efremov, R. G. & Sazanov, L. A. The coupling mechanism of respiratory complex I – a structural and evolutionary perspective. Biochim. Biophys. Acta 1817, 1785–1795 (2012)
Efremov, R. G., Baradaran, R. & Sazanov, L. A. The architecture of respiratory complex I. Nature 465, 441–445 (2010)
Efremov, R. G. & Sazanov, L. A. Respiratory complex I: ‘steam engine’ of the cell? Curr. Opin. Struct. Biol. 21, 532–540 (2011)
Angerer, H. et al. A scaffold of accessory subunits links the peripheral arm and the distal proton-pumping module of mitochondrial complex I. Biochem. J. 437, 279–288 (2011)
Hirst, J., Carroll, J., Fearnley, I. M., Shannon, R. J. & Walker, J. E. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim. Biophys. Acta 1604, 135–150 (2003)
Althoff, T., Mills, D. J., Popot, J. L. & Kuhlbrandt, W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1 . EMBO J. 30, 4652–4664 (2011)
Sazanov, L. A. & Hinchliffe, P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311, 1430–1436 (2006)
Berrisford, J. M. & Sazanov, L. A. Structural basis for the mechanism of respiratory complex I. J. Biol. Chem. 284, 29773–29783 (2009)
Hunte, C., Zickermann, V. & Brandt, U. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science 329, 448–451 (2010)
Efremov, R. G. & Sazanov, L. A. Structure of the membrane domain of respiratory complex I. Nature 476, 414–420 (2011)
Fearnley, I. M. & Walker, J. E. Conservation of sequences of subunits of mitochondrial complex I and their relationships with other proteins. Biochim. Biophys. Acta 1140, 105–134 (1992)
Mathiesen, C. & Hagerhall, C. Transmembrane topology of the NuoL, M and N subunits of NADH:quinone oxidoreductase and their homologues among membrane-bound hydrogenases and bona fide antiporters. Biochim. Biophys. Acta 1556, 121–132 (2002)
Sekiguchi, K., Murai, M. & Miyoshi, H. Exploring the binding site of acetogenin in the ND1 subunit of bovine mitochondrial complex I. Biochim. Biophys. Acta 1787, 1106–1111 (2009)
Nouws, J., Nijtmans, L. G., Smeitink, J. A. & Vogel, R. O. Assembly factors as a new class of disease genes for mitochondrial complex I deficiency: cause, pathology and treatment options. Brain 135, 12–22 (2012)
Angerer, H. et al. Tracing the tail of ubiquinone in mitochondrial complex I. Biochim. Biophys. Acta 1817, 1776–1784 (2012)
Wikström, M. & Hummer, G. Stoichiometry of proton translocation by respiratory complex I and its mechanistic implications. Proc. Natl Acad. Sci. USA 109, 4431–4436 (2012)
Verkhovsky, M., Bloch, D. A. & Verkhovskaya, M. Tightly-bound ubiquinone in the Escherichia coli respiratory Complex I. Biochim. Biophys. Acta 1817, 1550–1556 (2012)
Page, C. C., Moser, C. C., Chen, X. & Dutton, P. L. Natural engineering principles of electron tunnelling in biological oxidation–reduction. Nature 402, 47–52 (1999)
Kashani-Poor, N., Zwicker, K., Kerscher, S. & Brandt, U. A central functional role for the 49-kDa subunit within the catalytic core of mitochondrial complex I. J. Biol. Chem. 276, 24082–24087 (2001)
Ohnishi, T., Nakamaru-Ogiso, E. & Ohnishi, S. T. A new hypothesis on the simultaneous direct and indirect proton pump mechanisms in NADH-quinone oxidoreductase (complex I). FEBS Lett. 584, 4131–4137 (2010)
Berrisford, J. M., Thompson, C. J. & Sazanov, L. A. Chemical and NADH-induced, ROS-dependent, cross-linking between subunits of complex I from Escherichia coli and Thermus thermophilus. Biochemistry 47, 10262–10270 (2008)
Bai, F. et al. Conformational spread as a mechanism for cooperativity in the bacterial flagellar switch. Science 327, 685–689 (2010)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Brunger, A. T. Version 1.2 of the Crystallography and NMR system. Nature Protocols 2, 2728–2733 (2007)
Zhang, L. & Hermans, J. Hydrophilicity of cavities in proteins. Proteins 24, 433–438 (1996)
Huoponen, K., Vilkki, J., Aula, P., Nikoskelainen, E. K. & Savontaus, M. L. A new mtDNA mutation associated with Leber hereditary optic neuroretinopathy. Am. J. Hum. Genet. 48, 1147–1153 (1991)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D 67, 271–281 (2011)
The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Evans, P. R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D 67, 282–292 (2011)
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 8060–8065 (2006)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Krivov, G. G., Shapovalov, M. V. & Dunbrack, R. L., Jr Improved prediction of protein side-chain conformations with SCWRL4. Proteins 77, 778–795 (2009)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994)
Ashkenazy, H., Erez, E., Martz, E., Pupko, T. & Ben-Tal, N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 (2010)
Acknowledgements
This work was funded by the Medical Research Council. We thank the European Synchrotron Radiation Facility (ESRF) and the Swiss Light Source (SLS) for provision of synchrotron radiation facilities. We are grateful to the staff of beamlines ID29, ID23-2 (ESRF) and X06SA (SLS) for assistance.
Author information
Authors and Affiliations
Contributions
R.B. purified and crystallized the intact complex; J.M.B. purified and crystallized the membrane domain; G.S.M. performed co-crystallization and soaks with quinone analogues; all authors collected and analysed X-ray data; L.A.S. designed and supervised the project, analysed data and wrote the manuscript, with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion, Supplementary Tables 1-9, Supplementary Figures 1-7 and Supplementary References. (PDF 8477 kb)
Rights and permissions
About this article
Cite this article
Baradaran, R., Berrisford, J., Minhas, G. et al. Crystal structure of the entire respiratory complex I. Nature 494, 443–448 (2013). https://doi.org/10.1038/nature11871
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11871
This article is cited by
-
Disordered-to-ordered transitions in assembly factors allow the complex II catalytic subunit to switch binding partners
Nature Communications (2024)
-
Assembly and phylogeographical analysis of novel Taenia solium mitochondrial genomes suggest stratification within the African-American genotype
Parasites & Vectors (2023)
-
Human frataxin, the Friedreich ataxia deficient protein, interacts with mitochondrial respiratory chain
Cell Death & Disease (2023)
-
Mutation at the entrance of the quinone cavity severely disrupts quinone binding in respiratory complex I
Scientific Reports (2023)
-
H2O2 selectively damages the binuclear iron-sulfur cluster N1b of respiratory complex I
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.