Abstract
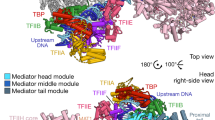
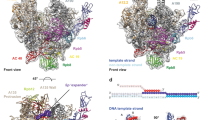
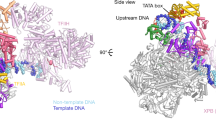
Gene transcription by RNA polymerase (Pol) II requires the coactivator complex Mediator. Mediator connects transcriptional regulators and Pol II, and is linked to human disease1,2,3,4. Mediator from the yeast Saccharomyces cerevisiae has a molecular mass of 1.4 megadaltons and comprises 25 subunits that form the head, middle, tail and kinase modules5,6,7. The head module constitutes one-half of the essential Mediator core8, and comprises the conserved9 subunits Med6, Med8, Med11, Med17, Med18, Med20 and Med22. Recent X-ray analysis of the S. cerevisiae head module at 4.3 Å resolution led to a partial architectural model with three submodules called neck, fixed jaw and moveable jaw10. Here we determine de novo the crystal structure of the head module from the fission yeast Schizosaccharomyces pombe at 3.4 Å resolution. Structure solution was enabled by new structures of Med6 and the fixed jaw, and previous structures of the moveable jaw11 and part of the neck12, and required deletion of Med20. The S. pombe head module resembles the head of a crocodile with eight distinct elements, of which at least four are mobile. The fixed jaw comprises tooth and nose domains, whereas the neck submodule contains a helical spine and one limb, with shoulder, arm and finger elements. The arm and the essential shoulder contact other parts of Mediator. The jaws and a central joint are implicated in interactions with Pol II and its carboxy-terminal domain, and the joint is required for transcription in vitro. The S. pombe head module structure leads to a revised model of the S. cerevisiae module, reveals a high conservation and flexibility, explains known mutations, and provides the basis for unravelling a central mechanism of gene regulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Protein Data Bank
Data deposits
Atomic coordinate files and structure factorswere deposited in the Protein Data Bank under accession codes 4H61 (Sp Med6), 4H62 (Sc Med17C–Med11C–Med22C) and 4H63 (Sp head module). See Supplementary Data for the coordinate file for the revised Sc head module model.
References
Conaway, R. C. & Conaway, J. W. Origins and activity of the Mediator complex. Semin. Cell Dev. Biol. 22, 729–734 (2011)
Chen, W. & Roeder, R. G. Mediator-dependent nuclear receptor function. Semin. Cell Dev. Biol. (2011)
Kaufmann, R. et al. Infantile cerebral and cerebellar atrophy is associated with a mutation in the MED17 subunit of the transcription preinitiation mediator complex. Am. J. Hum. Genet. 87, 667–670 (2010)
Myers, L. C. et al. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 12, 45–54 (1998)
Asturias, F. J., Jiang, Y. W., Myers, L. C., Gustafsson, C. M. & Kornberg, R. D. Conserved structures of mediator and RNA polymerase II holoenzyme. Science 283, 985–987 (1999)
Kang, J. S. et al. The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 276, 42003–42010 (2001)
Davis, J. A., Takagi, Y., Kornberg, R. & Asturias, F. Structure of the yeast RNA polymerase II holoenzyme: mediator conformation and polymerase interaction. Mol. Cell 10, 409–415 (2002)
Liu, Y., Ranish, J. A., Aebersold, R. & Hahn, S. Yeast nuclear extract contains two major forms of RNA polymerase II mediator complexes. J. Biol. Chem. 276, 7169–7175 (2001)
Bourbon, H. M. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36, 3993–4008 (2008)
Imasaki, T. et al. Architecture of the Mediator head module. Nature 475, 240–243 (2011)
Larivière, L. et al. Structure and TBP binding of the Mediator head subcomplex Med8–Med18–Med20. Nature Struct. Mol. Biol. 13, 895–901 (2006)
Seizl, M., Lariviere, L., Pfaffeneder, T., Wenzeck, L. & Cramer, P. Mediator head subcomplex Med11/22 contains a common helix bundle building block with a specific function in transcription initiation complex stabilization. Nucleic Acids Res. 39, 6291–6304 (2011)
Larivière, L., Seizl, M. & Cramer, P. A structural perspective on Mediator function. Curr. Opin. Cell Biol. (2012)
Larivière, L. et al. Structure-system correlation identifies a gene regulatory Mediator submodule. Genes Dev. 22, 872–877 (2008)
Baumli, S., Hoeppner, S. & Cramer, P. A conserved mediator hinge revealed in the structure of the MED7·MED21 (Med7·Srb7) heterodimer. J. Biol. Chem. 280, 18171–18178 (2005)
Guglielmi, B. et al. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 32, 5379–5391 (2004)
Cai, G. et al. Mediator head module structure and functional interactions. Nature Struct. Mol. Biol. 17, 273–279 (2010)
Takagi, Y. & Kornberg, R. D. Mediator as a general transcription factor. J. Biol. Chem. 281, 80–89 (2006)
Esnault, C. et al. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol. Cell 31, 337–346 (2008)
Lee, Y. C. & Kim, Y. J. Requirement for a functional interaction between mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol. Cell. Biol. 18, 5364–5370 (1998)
Lee, T. I. et al. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol. Cell. Biol. 18, 4455–4462 (1998)
Soutourina, J., Wydau, S., Ambroise, Y., Boschiero, C. & Werner, M. Direct interaction of RNA polymerase II and mediator required for transcription in vivo . Science 331, 1451–1454 (2011)
Tan, Q., Linask, K. L., Ebright, R. H. & Woychik, N. A. Activation mutants in yeast RNA polymerase II subunit RPB3 provide evidence for a structurally conserved surface required for activation in eukaryotes and bacteria. Genes Dev. 14, 339–348 (2000)
Thompson, C. M., Koleske, A. J., Chao, D. M. & Young, R. A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell 73, 1361–1375 (1993)
Näär, A. M., Taatjes, D. J., Zhai, W., Nogales, E. & Tjian, R. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev. 16, 1339–1344 (2002)
Nonet, M. L. & Young, R. A. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics 123, 715–724 (1989)
Koleske, A. J., Buratowski, S., Nonet, M. & Young, R. A. A novel transcription factor reveals a functional link between the RNA polymerase II CTD and TFIID. Cell 69, 883–894 (1992)
Takagi, Y. et al. Head module control of mediator interactions. Mol. Cell 23, 355–364 (2006)
Baek, H. J., Kang, Y. K. & Roeder, R. G. Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J. Biol. Chem. 281, 15172–15181 (2006)
Ranish, J. A., Yudkovsky, N. & Hahn, S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 13, 49–63 (1999)
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004)
Gouet, P., Courcelle, E., Stuart, D. I. & Metoz, F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 (1999)
Budisa, N. et al. High-level biosynthetic substitution of methionine in proteins by its analogs 2-aminohexanoic acid, selenomethionine, telluromethionine and ethionine in Escherichia coli . Eu. J. Biochem. 230, 788–796 (1995)
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Karplus, P. A. & Diederichs, K. Linking crystallographic model and data quality. Science 336, 1030–1033 (2012)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008)
Seizl, M. et al. A conserved GA element in TATA-less RNA polymerase II promoters. PLoS ONE 6, e27595 (2011)
Schneider, G. & Lindqvist, Y. Ta6Br14 is a useful cluster compound for isomorphous replacement in protein crystallography. Acta Crystallogr. D 50, 186–191 (1994)
Knäblein, J. et al. Ta6Br12 2+, a tool for phase determination of large biological assemblies by X-ray crystallography. J. Mol. Biol. 270, 1–7 (1997)
Cramer, P. et al. Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288, 640–649 (2000)
Girard, E., Stelter, M., Vicat, J. & Kahn, R. A new class of lanthanide complexes to obtain high-phasing-power heavy-atom derivatives for macromolecular crystallography. Acta Crystallogr. D 59, 1914–1922 (2003)
Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 (2007)
Terwilliger, T. C. Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972 (2000)
Blanc, E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol 60, 2210–2221 (2004)
Eswar, N. et al. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics Chapter 5, 5.6.1 (2006)
Acknowledgements
We thank S. Baumli, A. C. Cheung, A. Imhof and C. Schmidt for help. We acknowledge the crystallization facility at the Max Planck Institute of Biochemistry, Martinsried. Diffraction data were collected at the Swiss Light Source, Villigen, Switzerland. M.S. was supported by a Boehringer Ingelheim fellowship and the Elite Network of Bavaria. P.C. was supported by the Deutsche Forschungsgemeinschaft, SFB646, TR5, GraKo1721, SFB960, CIPSM, NIM, an Advanced Grant of the European Research Council, the LMUinnovativ project Bioimaging Network, the Vallee Foundation, and the Jung-Stiftung.
Author information
Authors and Affiliations
Contributions
L.L. designed crystallization constructs, established expression and purification strategies for head modules, solved the Med6 and Med17C–Med11C–Med22C structures, carried out crystallographic data analysis for Sp head module structure determination, built the Sc head module model, generated yeast strains, and performed in vivo studies. C.P. optimized Sp head module purification, purified and crystallized the Sp head module variants, and prepared heavy metal derivatives. L.L. and C.P. collected diffraction data for the head module and carried out model building and structural analysis. M.S. contributed to establishing expression and purification strategies and carried out transcription assays. L.W. provided technical help. F.K. optimized transcription assays. P.C. initiated and supervised the project. P.C., L.L., C.P. and M.S. prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Notes 1-2, Supplementary Figures 1-12, Supplementary Tables 1-3 and Supplementary References. (PDF 10571 kb)
Supplementary Data
This file contains the coordinates in PDB format for the revised Sc Mediator head module model. Subunits Med6, Med8, Med11, Med17, Med18, Med20 and Med22 correspond to chains F, H, K, Q, R, T and V, respectively. This file was originally missing and was added online on 19 December 2012. (TXT 457 kb)
Rights and permissions
About this article
Cite this article
Larivière, L., Plaschka, C., Seizl, M. et al. Structure of the Mediator head module. Nature 492, 448–451 (2012). https://doi.org/10.1038/nature11670
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11670
This article is cited by
-
The Mediator complex as a master regulator of transcription by RNA polymerase II
Nature Reviews Molecular Cell Biology (2022)
-
Structure of the human Mediator–RNA polymerase II pre-initiation complex
Nature (2021)
-
Transcription regulation by the Mediator complex
Nature Reviews Molecular Cell Biology (2018)
-
Crystal structure of human Mediator subunit MED23
Nature Communications (2018)
-
Development of an in silico method for the identification of subcomplexes involved in the biogenesis of multiprotein complexes in Saccharomyces cerevisiae
BMC Systems Biology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.