Abstract
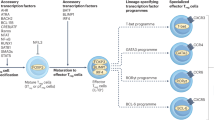
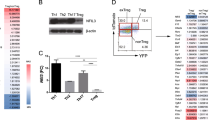
Regulatory T (Treg) cells, characterized by expression of the transcription factor forkhead box P3 (Foxp3), maintain immune homeostasis by suppressing self-destructive immune responses1,2,3,4. Foxp3 operates as a late-acting differentiation factor controlling Treg cell homeostasis and function5, whereas the early Treg-cell-lineage commitment is regulated by the Akt kinase and the forkhead box O (Foxo) family of transcription factors6,7,8,9,10. However, whether Foxo proteins act beyond the Treg-cell-commitment stage to control Treg cell homeostasis and function remains largely unexplored. Here we show that Foxo1 is a pivotal regulator of Tregcell function. Treg cells express high amounts of Foxo1 and display reduced T-cell-receptor-induced Akt activation, Foxo1 phosphorylation and Foxo1 nuclear exclusion. Mice with Treg-cell-specific deletion of Foxo1 develop a fatal inflammatory disorder similar in severity to that seen in Foxp3-deficient mice, but without the loss of Treg cells. Genome-wide analysis of Foxo1 binding sites reveals ∼300 Foxo1-bound target genes, including the pro-inflammatory cytokine Ifng, that do not seem to be directly regulated by Foxp3. These findings show that the evolutionarily ancient Akt–Foxo1 signalling module controls a novel genetic program indispensable for Treg cell function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sakaguchi, S., Yamaguchi, T., Nomura, T. & Ono, M. Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008)
Tang, Q. & Bluestone, J. A. The Foxp3+ regulatory T cell: a jack of all trades, master of regulation. Nature Immunol. 9, 239–244 (2008)
Feuerer, M., Hill, J. A., Mathis, D. & Benoist, C. Foxp3+ regulatory T cells: differentiation, specification, subphenotypes. Nature Immunol. 10, 689–695 (2009)
Shevach, E. M. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity 30, 636–645 (2009)
Rudensky, A. Y. Regulatory T cells and Foxp3. Immunol. Rev. 241, 260–268 (2011)
Haxhinasto, S., Mathis, D. & Benoist, C. The AKT–mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J. Exp. Med. 205, 565–574 (2008)
Sauer, S. et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc. Natl Acad. Sci. USA 105, 7797–7802 (2008)
Ouyang, W. et al. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nature Immunol. 11, 618–627 (2010)
Harada, Y. et al. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. J. Exp. Med. 207, 1381–1391 (2010)
Kerdiles, Y. M. et al. Foxo transcription factors control regulatory T cell development and function. Immunity 33, 890–904 (2010)
Ouyang, W., Beckett, O., Flavell, R. A. & Li, M. O. An essential role of the forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity 30, 358–371 (2009)
Kerdiles, Y. M. et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nature Immunol. 10, 176–184 (2009)
Patterson, S. J. et al. Cutting edge: PHLPP regulates the development, function, and molecular signaling pathways of regulatory T cells. J. Immunol. 186, 5533–5537 (2011)
Rubtsov, Y. P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008)
Kim, J. M., Rasmussen, J. P. & Rudensky, A. Y. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nature Immunol. 8, 191–197 (2007)
Lahl, K. et al. Selective depletion of Foxp3+ regulatory T cells induces a scurfy-like disease. J. Exp. Med. 204, 57–63 (2007)
Zhao, Y. et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nature Cell Biol. 12, 665–675 (2010)
Zhang, X. et al. Phosphorylation of serine 256 suppresses transactivation by FKHR (FOXO1) by multiple mechanisms. Direct and indirect effects on nuclear/cytoplasmic shuttling and DNA binding. J. Biol. Chem. 277, 45276–45284 (2002)
Sandri, M. et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399–412 (2004)
Gilley, J., Coffer, P. J. & Ham, J. FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J. Cell Biol. 162, 613–622 (2003)
Zheng, Y. et al. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 445, 936–940 (2007)
Marson, A. et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature 445, 931–935 (2007)
Gavin, M. A. et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature 445, 771–775 (2007)
Wing, K. et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science 322, 271–275 (2008)
Medina, I. et al. Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res. 38, W210–W213 (2010)
Hatton, R. D. et al. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity 25, 717–729 (2006)
Crellin, N. K., Garcia, R. V. & Levings, M. K. Altered activation of AKT is required for the suppressive function of human CD4+CD25+ T regulatory cells. Blood 109, 2014–2022 (2007)
Oldenhove, G. et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity 31, 772–786 (2009)
Dominguez-Villar, M., Baecher-Allan, C. M. & Hafler, D. A. Identification of T helper type 1-like, Foxp3+ regulatory T cells in human autoimmune disease. Nature Med. 17, 673–675 (2011)
McClymont, S. A. et al. Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J. Immunol. 186, 3918–3926 (2011)
Wan, Y. Y. & Flavell, R. A. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc. Natl Acad. Sci. USA 102, 5126–5131 (2005)
Driegen, S. et al. A generic tool for biotinylation of tagged proteins in transgenic mice. Transgenic Res. 14, 477–482 (2005)
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008)
Karolchik, D. et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496 (2004)
Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005)
Portales-Casamar, E. et al. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 38, D105–D110 (2010)
Matys, V. et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31, 374–378 (2003)
Smith, A. D., Sumazin, P., Xuan, Z. & Zhang, M. Q. DNA motifs in human and mouse proximal promoters predict tissue-specific expression. Proc. Natl Acad. Sci. USA 103, 6275–6280 (2006)
Li, L. GADEM: a genetic algorithm guided formation of spaced dyads coupled with an EM algorithm for motif discovery. J. Comput. Biol. 16, 317–329 (2009)
Gupta, S., Stamatoyannopoulos, J. A., Bailey, T. L. & Noble, W. S. Quantifying similarity between motifs. Genome Biol. 8, R24 (2007)
Irizarry, R. A. et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15 (2003)
López-Romero, P. Pre-processing and differential expression analysis of Agilent microRNA arrays using the AgiMicroRna Bioconductor library. BMC Genomics 12, 64 (2011)
Al-Shahrour, F., Diaz-Uriarte, R. & Dopazo, J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 20, 578–580 (2004)
Acknowledgements
We thank R. Flavell for the Foxp3-IRES-RFP mouse strain, K. Rajewsky and C. Xiao for the Rosa26 targeting vector and T. Unterman for the HA-hFoxo1(AAA) construct. This work was supported by the Starr Cancer Consortium (13-A123 to M.O.L., M.Q.Z. and G.A.), the Rita Allen Foundation (M.O.L.), National Bio Resource Project (NBRPC) (2012CB316503 to M.Q.Z.) and National Institutes of Health (HG001696 to M.Q.Z.).
Author information
Authors and Affiliations
Contributions
W.O., W.L., C.T.L., N.Y., M.H., G.A., M.Q.Z. and M.O.L. designed the research and analysed the data; W.O., W.L., C.T.L., N.Y., M.H., M.V.K., M.P., P.C., Q.M. and Y.M. did the experiments; D.M. and A.Y.R. provided birA and Foxp3cre mouse strains; K.Z. supervised the ChIP-seq experiment; and W.O., W.L. and M.O.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-14. (PDF 5377 kb)
Supplementary Table 1
This file contains a complete list of candidate binding sites detected in antibody and biotin Foxo1 ChIP-Seq (overlapping peaks have been merged). (XLSX 253 kb)
Supplementary Table 2
This file shows differentially expressed genes in Foxo1-deficient Tregs (1.5X or greater). (XLSX 232 kb)
Supplementary Table 3
This file contains a complete list of candidate binding sites detected in antibody and biotin Foxo1 ChIP-Seq (selecting for peaks nearby genes that are differentially expressed in Foxo1 KO and rescued by Foxo1AAA). (XLSX 91 kb)
Supplementary Table 4
This file shows gene ontology analysis of Foxo1 direct target genes. (XLSX 47 kb)
Foxo1 nuclear exclusion in response to high-dose CD3 antibody
CD4+ cells were isolated from Foxo1tag/tag Foxp3-IRES-RFP mice, and stimulated with plate-bound anti-CD3 (0.1 μg/ml) and anti-CD28 (1.0 μg/ml). GFP, RFP and bright-field images were acquired every 2 min after addition of cells using a fluorescence videomicroscope. (MOV 68 kb)
Foxo1 nuclear exclusion in response to low-dose CD3 antibody
CD4+ cells were isolated from Foxo1tag/tag Foxp3-IRES-RFP mice, and stimulated with plate-bound anti-CD3 (0.01 μg/ml) and anti-CD28 (1.0 μg/ml). GFP, RFP and bright-field images were acquired every 2 min after addition of cells using a fluorescence videomicroscope (MOV 110 kb)
Rights and permissions
About this article
Cite this article
Ouyang, W., Liao, W., Luo, C. et al. Novel Foxo1-dependent transcriptional programs control Treg cell function. Nature 491, 554–559 (2012). https://doi.org/10.1038/nature11581
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11581
This article is cited by
-
The regulation and differentiation of regulatory T cells and their dysfunction in autoimmune diseases
Nature Reviews Immunology (2024)
-
Type 1 interferons and Foxo1 down-regulation play a key role in age-related T-cell exhaustion in mice
Nature Communications (2024)
-
How autoreactive thymocytes differentiate into regulatory versus effector CD4+ T cells after avoiding clonal deletion
Nature Immunology (2023)
-
T helper cell polarity determines salt sensitivity and hypertension development
Hypertension Research (2023)
-
IL-9 aggravates SARS-CoV-2 infection and exacerbates associated airway inflammation
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.