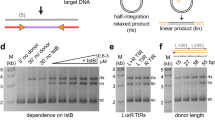
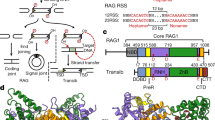
Abstract
Studies of bacteriophage Mu transposition paved the way for understanding retroviral integration and V(D)J recombination as well as many other DNA transposition reactions. Here we report the structure of the Mu transpososome—Mu transposase (MuA) in complex with bacteriophage DNA ends and target DNA—determined from data that extend anisotropically to 5.2 Å, 5.2 Å and 3.7 Å resolution, in conjunction with previously determined structures of individual domains. The highly intertwined structure illustrates why chemical activity depends on formation of the synaptic complex, and reveals that individual domains have different roles when bound to different sites. The structure also provides explanations for the increased stability of the final product complex and for its preferential recognition by the ATP-dependent unfoldase ClpX. Although MuA and many other recombinases share a structurally conserved ‘DDE’ catalytic domain, comparisons among the limited set of available complex structures indicate that some conserved features, such as catalysis in trans and target DNA bending, arose through convergent evolution because they are important for function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Yanagihara, K. & Mizuuchi, K. Mismatch-targeted transposition of Mu: a new strategy to map genetic polymorphism. Proc. Natl Acad. Sci. USA 99, 11317–11321 (2002)
Haapa, S., Taira, S., Heikkinen, E. & Savilahti, H. An efficient and accurate integration of mini-Mu transposons in vitro: a general methodology for functional genetic analysis and molecular biology applications. Nucleic Acids Res. 27, 2777–2784 (1999)
Mizuuchi, K. In vitro transposition of bacteriophage Mu: a biochemical approach to a novel replication reaction. Cell 35, 785–794 (1983)
Montaño, S. P. & Rice, P. A. Moving DNA around: DNA transposition and retroviral integration. Curr. Opin. Struct. Biol. 21, 370–378 (2011)
Davies, D. R., Goryshin, I. Y., Reznikoff, W. S. & Rayment, I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289, 77–85 (2000)
Maertens, G. N., Hare, S. & Cherepanov, P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 468, 326–329 (2010)
Richardson, J. M., Colloms, S. D., Finnegan, D. J. & Walkinshaw, M. D. Molecular architecture of the Mos1 paired-end complex: the structural basis of DNA transposition in a eukaryote. Cell 138, 1096–1108 (2009)
Choi, W. & Harshey, R. M. DNA repair by the cryptic endonuclease activity of Mu transposase. Proc. Natl Acad. Sci. USA 107, 10014–10019 (2010)
Chaconas, G., Kennedy, D. L. & Evans, D. Predominant integration end products of infecting bacteriophage Mu DNA are simple insertions with no preference for integration of either Mu DNA strand. Virology 128, 48–59 (1983)
Lavoie, B. D., Chan, B. S., Allison, R. G. & Chaconas, G. Structural aspects of a higher order nucleoprotein complex: induction of an altered DNA structure at the Mu-host junction of the Mu type 1 transpososome. EMBO J. 10, 3051–3059 (1991)
Surette, M. G., Buch, S. J. & Chaconas, G. Transpososomes: stable protein-DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell 49, 253–262 (1987)
Au, T. K., Pathania, S. & Harshey, R. M. True reversal of Mu integration. EMBO J. 23, 3408–3420 (2004)
Mizuuchi, M., Rice, P. A., Wardle, S. J., Haniford, D. B. & Mizuuchi, K. Control of transposase activity within a transpososome by the configuration of the flanking DNA segment of the transposon. Proc. Natl Acad. Sci. USA 104, 14622–14627 (2007)
Kruklitis, R., Welty, D. J. & Nakai, H. ClpX protein of Escherichia coli activates bacteriophage Mu transposase in the strand transfer complex for initiation of Mu DNA synthesis. EMBO J. 15, 935–944 (1996)
Levchenko, I., Luo, L. & Baker, T. A. Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes Dev. 9, 2399–2408 (1995)
Mhammedi-Alaoul, A., Pato, M., Gama, M. J. & Toussaint, A. A new component of bacteriophage Mu replicative transposition machinery: the Escherichia coli ClpX protein. Mol. Microbiol. 11, 1109–1116 (1994)
Abdelhakim, A. H., Oakes, E. C., Sauer, R. T. & Baker, T. A. Unique contacts direct high-priority recognition of the tetrameric Mu transposase-DNA complex by the AAA+ unfoldase ClpX. Mol. Cell 30, 39–50 (2008)
Savilahti, H., Rice, P. A. & Mizuuchi, K. The phage Mu transpososome core: DNA requirements for assembly and function. EMBO J. 14, 4893–4903 (1995)
Baker, T. A. & Mizuuchi, K. DNA-promoted assembly of the active tetramer of the Mu transposase. Genes Dev. 6, 2221–2232 (1992)
Yuan, J. F., Beniac, D. R., Chaconas, G. & Ottensmeyer, F. P. 3D reconstruction of the Mu transposase and the Type 1 transpososome: a structural framework for Mu DNA transposition. Genes Dev. 19, 840–852 (2005)
Savilahti, H. & Mizuuchi, K. Mu transpositional recombination: donor DNA cleavage and strand transfer in trans by the Mu transposase. Cell 85, 271–280 (1996)
Aldaz, H., Schuster, E. & Baker, T. A. The interwoven architecture of the Mu transposase couples DNA synapsis to catalysis. Cell 85, 257–269 (1996)
Krementsova, E., Giffin, M. J., Pincus, D. & Baker, T. A. Mutational analysis of the Mu transposase. Contributions of two distinct regions of domain II to recombination. J. Biol. Chem. 273, 31358–31365 (1998)
Namgoong, S. Y., Sankaralingam, S. & Harshey, R. M. Altering the DNA-binding specificity of Mu transposase in vitro. Nucleic Acids Res. 26, 3521–3527 (1998)
Zou, A. H., Leung, P. C. & Harshey, R. M. Transposase contacts with mu DNA ends. J. Biol. Chem. 266, 20476–20482 (1991)
Tanaka, Y. et al. Crystal structure of the CENP-B protein-DNA complex: the DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA. EMBO J. 20, 6612–6618 (2001)
Watkins, S., van Pouderoyen, G. & Sixma, T. K. Structural analysis of the bipartite DNA-binding domain of Tc3 transposase bound to transposon DNA. Nucleic Acids Res. 32, 4306–4312 (2004)
Craigie, R., Mizuuchi, M. & Mizuuchi, K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell 39, 387–394 (1984)
Kuo, C. F., Zou, A. H., Jayaram, M., Getzoff, E. & Harshey, R. DNA-protein complexes during attachment-site synapsis in Mu DNA transposition. EMBO J. 10, 1585–1591 (1991)
Mizuuchi, M., Baker, T. A. & Mizuuchi, K. DNase protection analysis of the stable synaptic complexes involved in Mu transposition. Proc. Natl Acad. Sci. USA 88, 9031–9035 (1991)
Rice, P. & Mizuuchi, K. Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration. Cell 82, 209–220 (1995)
Wu, Z. & Chaconas, G. A novel DNA binding and nuclease activity in domain III of Mu transposase: evidence for a catalytic region involved in donor cleavage. EMBO J. 14, 3835–3843 (1995)
Abdelhakim, A. H., Sauer, R. T. & Baker, T. A. The AAA+ ClpX machine unfolds a keystone subunit to remodel the Mu transpososome. Proc. Natl Acad. Sci. USA 107, 2437–2442 (2010)
Burton, B. M. & Baker, T. A. Mu transpososome architecture ensures that unfolding by ClpX or proteolysis by ClpXP remodels but does not destroy the complex. Chem. Biol. 10, 463–472 (2003)
Naigamwalla, D. Z., Coros, C. J., Wu, Z. & Chaconas, G. Mutations in domain III α of the Mu transposase: evidence suggesting an active site component which interacts with the Mu-host junction. J. Mol. Biol. 282, 265–274 (1998)
Yang, J. Y., Kim, K., Jayaram, M. & Harshey, R. M. A domain sharing model for active site assembly within the Mu A tetramer during transposition: the enhancer may specify domain contributions. EMBO J. 14, 2374–2384 (1995)
Surette, M. G. & Chaconas, G. The Mu transpositional enhancer can function in trans: requirement of the enhancer for synapsis but not strand cleavage. Cell 68, 1101–1108 (1992)
Mizuuchi, M. & Mizuuchi, K. Conformational isomerization in phage Mu transpososome assembly: effects of the transpositional enhancer and of MuB. EMBO J. 20, 6927–6935 (2001)
Harshey, R. M. & Jayaram, M. The mu transpososome through a topological lens. Crit. Rev. Biochem. Mol. Biol. 41, 387–405 (2006)
Craigie, R. & Mizuuchi, K. Role of DNA topology in Mu transposition: mechanism of sensing the relative orientation of two DNA segments. Cell 45, 793–800 (1986)
Surette, M. G. & Chaconas, G. A protein factor which reduces the negative supercoiling requirement in the Mu DNA strand transfer reaction is Escherichia coli integration host factor. J. Biol. Chem. 264, 3028–3034 (1989)
Allison, R. G. & Chaconas, G. Role of the A protein-binding sites in the in vitro transposition of Mu DNA. A complex circuit of interactions involving the Mu ends and the transpositional enhancer. J. Biol. Chem. 267, 19963–19970 (1992)
Jiang, H., Yang, J. Y. & Harshey, R. M. Criss-crossed interactions between the enhancer and the att sites of phage Mu during DNA transposition. EMBO J. 18, 3845–3855 (1999)
Craig, N. L. Mobile DNA II (ASM Press, 2002)
Pribil, P. A. & Haniford, D. B. Target DNA bending is an important specificity determinant in target site selection in Tn10 transposition. J. Mol. Biol. 330, 247–259 (2003)
Swinger, K. K. & Rice, P. A. Structure-based analysis of HU-DNA binding. J. Mol. Biol. 365, 1005–1016 (2007)
Levchenko, I., Yamauchi, M. & Baker, T. A. ClpX and MuB interact with overlapping regions of Mu transposase: implications for control of the transposition pathway. Genes Dev. 11, 1561–1572 (1997)
Clubb, R. T., Schumacher, S., Mizuuchi, K., Gronenborn, A. M. & Clore, G. M. Solution structure of the Iγ subdomain of the Mu end DNA-binding domain of phage Mu transposase. J. Mol. Biol. 273, 19–25 (1997)
Schumacher, S. et al. Solution structure of the Mu end DNA-binding Iβ subdomain of phage Mu transposase: modular DNA recognition by two tethered domains. EMBO J. 16, 7532–7541 (1997)
Baker, T. A., Mizuuchi, M., Savilahti, H. & Mizuuchi, K. Division of labor among monomers within the Mu transposase tetramer. Cell 74, 723–733 (1993)
Ducruix, A. & Giegg, R. in Preparation of Selenomethionyl Protein Crystals (eds Dublie, S. & Carter, C. W. ) (Oxford Univ. Press, 1992)
Otwinowski, Z. & Minor, W. in Methods in Enzymology Vol. 276 (eds Carter, C. W. & Sweet, R. M. ) 307–326 (Academic, 1997)
Sheldrick, G. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008)
Brünger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
CCP4. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Zhang, K. Y., Cowtan, K. & Main, P. Combining constraints for electron-density modification. Methods Enzymol. 277, 53–64 (1997)
Emsley, P., Lohkamp, B., Scott, W. & Cowtan, K. Features and Development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Schröder, G. F., Levitt, M. & Brunger, A. T. Super-resolution biomolecular crystallography with low-resolution data. Nature 464, 1218–1222 (2010)
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 8060–8065 (2006)
Zheng, G., Lu, X. J. & Olson, W. K. Web 3DNA–a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 37, W240–W246 (2009)
Lavoie, B. D. & Chaconas, G. Site-specific HU binding in the Mu transpososome: conversion of a sequence-independent DNA-binding protein into a chemical nuclease. Genes Dev. 7, 2510–2519 (1993)
Swinger, K. K. & Rice, P. A. IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 14, 28–35 (2004)
Acknowledgements
We thank K. Mizuuchi for initiating this project, K. K. Swinger and B. Vertessy for early crystallization efforts, and X. Yang and the staff of APS beamlines 14, 19 and 21 for assistance with data collection. This work was funded in part by NIH grant GM086826 (to P.A.R.).
Author information
Authors and Affiliations
Contributions
S.P.M. carried out most of the crystallographic work, Y.Z.P. grew the first diffracting transpososome crystals and assisted with all other aspects of the project, and P.A.R. designed the project and assisted in computational work and interpretation of the results.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-4, Supplementary References and Supplementary Table 1. (PDF 1162 kb)
Ribbon drawing of the transpososome structure rotating 360°
The complex is rotating about the crystallographic twofold axis that relates the red and blue halves. Colours are as in the main text: bacteriophage Mu end DNAs are red and blue, target DNA black, and the scissile phosphate and active site residues are yellow. The darker-colored subunits catalyze DNA cleavage and strand transfer and the lighter-colored subunits aid in complex assembly and stability. (MPG 4785 kb)
Closeup view of the experimental electron density, after improvement with Parrot, and contoured at 1.3 and 2.3 Sigma, rotating 360°
The rotation axis and colors are as in the main text and Supplementary Video 1. (MPG 9535 kb)
Rights and permissions
About this article
Cite this article
Montaño, S., Pigli, Y. & Rice, P. The Mu transpososome structure sheds light on DDE recombinase evolution. Nature 491, 413–417 (2012). https://doi.org/10.1038/nature11602
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11602
This article is cited by
-
Zinc-finger BED domains drive the formation of the active Hermes transpososome by asymmetric DNA binding
Nature Communications (2023)
-
Structure reveals why genome folding is necessary for site-specific integration of foreign DNA into CRISPR arrays
Nature Structural & Molecular Biology (2023)
-
Structure and function of retroviral integrase
Nature Reviews Microbiology (2022)
-
Characterization and Genome Analysis of a Novel Mu-like Phage VW-6B Isolated from the Napahai Plateau Wetland of China
Current Microbiology (2021)
-
Deep sequencing reveals new roles for MuB in transposition immunity and target-capture, and redefines the insular Ter region of E. coli
Mobile DNA (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.