Abstract
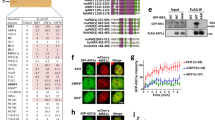
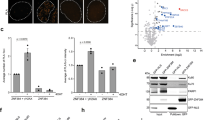
Several homology-dependent pathways can repair potentially lethal DNA double-strand breaks (DSBs). The first step common to all homologous recombination reactions is the 5′–3′ degradation of DSB ends that yields the 3′ single-stranded DNA required for the loading of checkpoint and recombination proteins. In yeast, the Mre11–Rad50–Xrs2 complex (Xrs2 is known as NBN or NBS1 in humans) and Sae2 (known as RBBP8 or CTIP in humans) initiate end resection, whereas long-range resection depends on the exonuclease Exo1, or the helicase–topoisomerase complex Sgs1–Top3–Rmi1 together with the endonuclease Dna2 (refs 1–6). DSBs occur in the context of chromatin, but how the resection machinery navigates through nucleosomal DNA is a process that is not well understood7. Here we show that the yeast Saccharomyces cerevisiae Fun30 protein and its human counterpart SMARCAD1 (ref. 8), two poorly characterized ATP-dependent chromatin remodellers of the Snf2 ATPase family, are directly involved in the DSB response. Fun30 physically associates with DSB ends and directly promotes both Exo1- and Sgs1-dependent end resection through a mechanism involving its ATPase activity. The function of Fun30 in resection facilitates the repair of camptothecin-induced DNA lesions, although it becomes dispensable when Exo1 is ectopically overexpressed. Interestingly, SMARCAD1 is also recruited to DSBs, and the kinetics of recruitment is similar to that of EXO1. The loss of SMARCAD1 impairs end resection and recombinational DNA repair, and renders cells hypersensitive to DNA damage resulting from camptothecin or poly(ADP-ribose) polymerase inhibitor treatments. These findings unveil an evolutionarily conserved role for the Fun30 and SMARCAD1 chromatin remodellers in controlling end resection, homologous recombination and genome stability in the context of chromatin.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
26 September 2012
Reference numbering in the online-only Methods section was corrected.
References
Mimitou, E. P. & Symington, L. S. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774 (2008)
Zhu, Z., Chung, W. H. S. h. i. m. E. Y., Lee, S. E. & Ira, G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994 (2008)
Gravel, S., Chapman, J. R., Magill, C. & Jackson, S. P. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22, 2767–2772 (2008)
Cejka, P. et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467, 112–116 (2010)
Nicolette, M. L. et al. Mre11–Rad50–Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nature Struct. Mol. Biol. 17, 1478–1485 (2010)
Niu, H. et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae . Nature 467, 108–111 (2010)
Sinha, M. & Peterson, C. L. Chromatin dynamics during repair of chromosomal DNA double-strand breaks. Epigenomics 1, 371–385 (2009)
Awad, S., Ryan, D., Prochasson, P., Owen-Hughes, T. & Hassan, A. H. The Snf2 homolog Fun30 acts as a homodimeric ATP-dependent chromatin-remodeling enzyme. J. Biol. Chem. 285, 9477–9484 (2010)
Neves-Costa, A., Will, W. R., Vetter, A. T., Miller, J. R. & Varga-Weisz, P. The SNF2-family member Fun30 promotes gene silencing in heterochromatic loci. PLoS ONE 4, e8111 (2009)
Ouspenski, I. I., Elledge, S. J. & Brinkley, B. R. New yeast genes important for chromosome integrity and segregation identified by dosage effects on genome stability. Nucleic Acids Res. 27, 3001–3008 (1999)
Marrero, V. A. & Symington, L. S. Extensive DNA end processing by Exo1 and Sgs1 inhibits break-induced replication. PLoS Genet. 6, e1001007 (2010)
White, C. I. & Haber, J. E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae . EMBO J. 9, 663–673 (1990)
Shim, E. Y. et al. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 29, 3370–3380 (2010)
Chen, C. C. e. t. a. l. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134, 231–243 (2008)
Clerici, M., Mantiero, D., Lucchini, G. & Longhese, M. P. The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep. 7, 212–218 (2006)
Zubko, M. K. Exo1 and Rad24 differentially regulate generation of ssDNA at telomeres of Saccharomyces cerevisiae cdc13–1 mutants. Genetics 168, 103–115 (2004)
Grandin, N. & Charbonneau, M. Control of the yeast telomeric senescence survival pathways of recombination by the Mec1 and Mec3 DNA damage sensors and RPA. Nucleic Acids Res. 35, 822–838 (2007)
Garvik, B., Carson, M. & Hartwell, L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15, 6128–6138 (1995)
Matsuoka, S. et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 (2007)
Beli, P. et al. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol. Cell 46, 212–225 (2012)
Tomimatsu, N. et al. Exo1 plays a major role in DNA end resection in humans and influences double-strand break repair and damage signaling decisions. DNA Repair 11, 441–448 (2012)
Bolderson, E. et al. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. 38, 1821–1831 (2010)
Strålfors, A., Walfridsson, J., Bhuiyan, H. & Ekwall, K. The FUN30 chromatin remodeler, Fft3, protects centromeric and subtelomeric domains from euchromatin formation. PLoS Genet. 7, e1001334 (2011)
Rowbotham, S. P. et al. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol. Cell 42, 285–296 (2011)
Ooi, S. L., Shoemaker, D. D. & Boeke, J. D. DNA microarray-based genetic screen for nonhomologous end-joining mutants in Saccharomyces cerevisiae . Science 294, 2552–2556 (2001)
Bärtsch, S., Kang, L. E. & Symington, L. S. RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol. Cell. Biol. 20, 1194–1205 (2000)
van Attikum, H., Fritsch, O. & Gasser, S. M. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 26, 4113–4125 (2007)
Tomimatsu, N., Mukherjee, B. & Burma, S. Distinct roles of ATR and DNA-PKcs in triggering DNA damage responses in ATM-deficient cells. EMBO Rep. 10, 629–635 (2009)
Shanbhag, N. M., Rafalska-Metcalf, I. U., Balane-Bolivar, C., Janicki, S. M. & Greenberg, R. A. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141, 970–981 (2010)
Janke, C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004)
Pardo, B., Ma, E. & Marcand, S. Mismatch tolerance by DNA polymerase Pol4 in the course of nonhomologous end joining in Saccharomyces cerevisiae . Genetics 172, 2689–2694 (2006)
Moreau, S., Morgan, E. A. & Symington, L. S. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 159, 1423–1433 (2001)
Burke, D. & Strathern, J. in Methods in Yeast Genetics: a Cold Spring Harbor Laboratory Course Manual (eds Amberg, D. C., Burke, D. & Strathern, J. N. ). (2005)
Herskowitz, I. & Jensen, R. E. Putting the HO gene to work: practical uses for mating-type switching. Methods Enzymol. 194, 132–146 (1991)
Decourty, L. et al. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc. Natl Acad. Sci. USA 105, 5821–5826 (2008)
Martini, E. M. D., Keeney, S. & Osley, M. A. A role for histone H2B during repair of UV-induced DNA damage in Saccharomyces cerevisiae . Genetics 160, 1375–1387 (2002)
Bekker-Jensen, S. et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 173, 195–206 (2006)
Sporbert, A., Gahl, A., Ankerhold, R., Leonhardt, H. & Cardoso, M. C. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol. Cell 10, 1355–1365 (2002)
Acknowledgements
We thank G. Ira for sharing unpublished data and S. Janicki, R. Greenberg, L. Symington and all the laboratories from the Centre National de la Recherche Scientifique (CNRS) UPR3081 for providing reagents. We thank S. Coulon for help in the analysis of the fun30 repressible allele, I. Lafontaine for support in statistical analyses, C. V. Camacho for generating the V5-EXO1 constructs and A. Guénolé, R. Srivas, T. Ideker, K. Vreeken and M. Vermeulen for help in searching for Fun30 interactors. B.L. is grateful to B. Dujon for hosting him and providing the opportunity to perform the BIR screen. S.B. is supported by grants from the National Institutes of Health (RO1 CA149461), National Aeronautics and Space Administration (NNX10AE08G) and the Cancer Prevention and Research Institute of Texas (RP100644). H.v.A. receives funding from the Netherlands Organization for Scientific Research (NWO-VIDI grant) and Human Frontiers Science Program (HFSP-CDA grant). B.L. is supported by grants from the CNRS (ATIP) and the Agence Nationale de la Recherche (ANR-10-BLAN-1606-03).
Author information
Authors and Affiliations
Contributions
B.L. and A.T. performed the genetic screen and B.L. identified the resection defect of fun30Δ. T.C. constructed yeast strains and plasmids and performed the yeast ChIP experiments. R.L. constructed yeast strains and performed ssDNA analysis by alkaline gels, BIR and gap-repair assays. R.L. and T.C. analysed SSA defects. N.T. and B.M. performed all of the SMARCAD1 knockdown experiments in human cells and the DR-GFP assays. E.M. designed and built the strain containing the inducible I-SceI cut site at HIS3, performed the micrococcal nuclease assay and contributed to data analysis. B.K. performed the analysis of survivors in the absence of telomerase. K.D. assisted R.L. and K.D., R.L. and T.C. performed fun30 DNA-damage-sensitivity assays. W.W.W. examined the localization of SMARCAD1 at FokI-induced DSBs. T.C., S.B., H.v.A. and B.L. designed the experiments and analysed the data. H.v.A. and B.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-13, the full legend for Supplementary Table 1, Supplementary Tables 2 and 3 and Supplementary References. This file was replaced on 26 September 2012 to correct the numbering of the references. (PDF 5558 kb)
Supplementary Data
This file contains Supplementary Table 1 (see Supplementary Information for full legend). (TXT 249 kb)
Rights and permissions
About this article
Cite this article
Costelloe, T., Louge, R., Tomimatsu, N. et al. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature 489, 581–584 (2012). https://doi.org/10.1038/nature11353
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11353
This article is cited by
-
Antler-derived microRNA PC-5p-1090 inhibits HCC cell proliferation, migration, and invasion by targeting MARCKS, SMARCAD1, and SOX9
Functional & Integrative Genomics (2023)
-
Smarcad1 mediates microbiota-induced inflammation in mouse and coordinates gene expression in the intestinal epithelium
Genome Biology (2020)
-
DNA damage response manages cell cycle restriction of senile multipotent mesenchymal stromal cells
Molecular Biology Reports (2020)
-
Quantitative sensing and signalling of single-stranded DNA during the DNA damage response
Nature Communications (2019)
-
1H, 13C and 15N resonance assignments for the tandem CUE domains from chromatin remodeler SMARCAD1
Biomolecular NMR Assignments (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.