Abstract
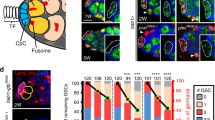
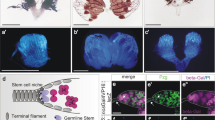
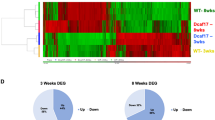
Adult stem cells support tissue homeostasis and repair throughout the life of an individual. During ageing, numerous intrinsic and extrinsic changes occur that result in altered stem-cell behaviour and reduced tissue maintenance and regeneration. In the Drosophila testis, ageing results in a marked decrease in the self-renewal factor Unpaired (Upd), leading to a concomitant loss of germline stem cells. Here we demonstrate that IGF-II messenger RNA binding protein (Imp) counteracts endogenous small interfering RNAs to stabilize upd (also known as os) RNA. However, similar to upd, Imp expression decreases in the hub cells of older males, which is due to the targeting of Imp by the heterochronic microRNA let-7. In the absence of Imp, upd mRNA therefore becomes unprotected and susceptible to degradation. Understanding the mechanistic basis for ageing-related changes in stem-cell behaviour will lead to the development of strategies to treat age-onset diseases and facilitate stem-cell-based therapies in older individuals.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Schofield, R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells 4, 7–25 (1978)
Jones, D. L. & Rando, T. A. Emerging models and paradigms for stem cell ageing. Nature Cell Biol. 13, 506–512 (2011)
Voog, J. & Jones, D. L. Stem cells and the Niche: a dynamic duo. Cell Stem Cell 6, 103–115 (2010)
Fuller, M. T. in The Development of Drosophila Melanogaster (eds Bate, M. & Martinez-Arias, A. ) 71–147 (Cold Spring Harbor Laboratory Press, 1993)
Boyle, M., Wong, C., Rocha, M. & Jones, D. L. Decline in self-renewal factors contributes to aging of the stem cell niche in the Drosophila testis. Cell Stem Cell 1, 470–478 (2007)
Buszczak, M. et al. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics 175, 1505–1531 (2007)
Fabrizio, J. J. et al. Imp (IGF-II mRNA-binding protein) is expressed during spermatogenesis in Drosophila melanogaster . Fly 2, 47–48 (2008)
Yisraeli, J. K. VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol. Cell 97, 87–96 (2005)
Brand, A. H., Manoukian, A. S. & Perrimon, N. Ectopic expression in Drosophila . Methods Cell Biol. 44, 635–654 (1994)
Munro, T. P., Kwon, S., Schnapp, B. J. & St Johnston, D. A repeated IMP-binding motif controls oskar mRNA translation and anchoring independently of Drosophila melanogaster IMP. J. Cell Biol. 172, 577–588 (2006)
Nabel-Rosen, H., Dorevitch, N., Reuveny, A. & Volk, T. The balance between two isoforms of the Drosophila RNA-binding protein how controls tendon cell differentiation. Mol. Cell 4, 573–584 (1999)
Hafner, M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 (2010)
Bhattacharyya, S. N., Habermacher, R., Martine, U., Closs, E. I. & Filipowicz, W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125, 1111–1124 (2006)
Elcheva, I., Goswami, S., Noubissi, F. K. & Spiegelman, V. S. CRD-BP protects the coding region of βTrCP1 mRNA from miR-183-mediated degradation. Mol. Cell 35, 240–246 (2009)
Czech, B. et al. An endogenous small interfering RNA pathway in Drosophila . Nature 453, 798–802 (2008)
Lee, Y. S. et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117, 69–81 (2004)
Carthew, R. W. & Sontheimer, E. J. Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655 (2009)
Golden, D. E., Gerbasi, V. R. & Sontheimer, E. J. An inside job for siRNAs. Mol. Cell 31, 309–312 (2008)
Czech, B. et al. Hierarchical rules for Argonaute loading in Drosophila . Mol. Cell 36, 445–456 (2009)
Geng, C. & Macdonald, P. M. Imp associates with squid and Hrp48 and contributes to localized expression of gurken in the oocyte. Mol. Cell. Biol. 26, 9508–9516 (2006)
Wang, L. & Jones, D. L. The effects of aging on stem cell behavior in Drosophila . Exp. Gerontol. 46, 340–344 (2010)
Boyerinas, B. et al. Identification of let-7-regulated oncofetal genes. Cancer Res. 68, 2587–2591 (2008)
Nishino, J., Kim, I., Chada, K. & Morrison, S. J. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell 135, 227–239 (2008)
Zhao, C. et al. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc. Natl Acad. Sci. USA 107, 1876–1881 (2010)
Landgraf, P. et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414 (2007)
Chen, C. et al. Defining embryonic stem cell identity using differentiation-related microRNAs and their potential targets. Mamm. Genome 18, 316–327 (2007)
Rybak, A., Fuchs, H., Smirnova, L., Brandt, C., Pohl, E. E., Nitsch, R. & Wulczyn, F. G. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nature Cell Biol. 10, 987–993 (2008)
Melton, C., Judson, R. L. & Blelloch, R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463, 621–626 (2010)
Iliopoulos, D., Hirsch, H. A. & Struhl, K. An epigenetic switch involving NF-κB, Lin28, let-7 microRNA, and IL6 links inflammation to cell transformation. Cell 139, 693–706 (2009)
Boyerinas, B., Park, S. M., Hau, A., Murmann, A. E. & Peter, M. E. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer 17, F19–F36 (2010)
Zhu, H. et al. The Lin28/let-7 axis regulates glucose metabolism. Cell 147, 81–94 (2011)
Li, X., Cassidy, J. J., Reinke, C. A., Fischboeck, S. & Carthew, R. W. A microRNA imparts robustness against environmental fluctuation during development. Cell 137, 273–282 (2009)
Boylan, K. L. et al. Motility screen identifies Drosophila IGF-II mRNA-binding protein–zipcode-binding protein acting in oogenesis and synaptogenesis. PLoS Genet. 4, e36 (2008)
Kitadate, Y. et al. Boss/Sev signaling from germline to soma restricts germline-stem-cell-niche formation in the anterior region of Drosophila male gonads. Dev. Cell 13, 151–159 (2007)
Sokol, N. S. et al. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 22, 1591–1596 (2008)
Caldwell, J. C., Fineberg, S. K. & Eberl, D. F. reduced ocelli encodes the leucine rich repeat protein Pray For Elves in Drosophila melanogaster . Fly 1, 146–152 (2007)
Hime, G. R., Brill, J. A. & Fuller, M. T. Assembly of ring canals in the male germ line from structural components of the contractile ring. J. Cell Sci. 109, 2779–2788 (1996)
Harrison, D. A., McCoon, P. E., Binari, R., Gilman, M. & Perrimon, N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 12, 3252–3263 (1998)
Bailey, T. L. & Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 (1994)
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C T method. Nature Protocols 3, 1101–1108 (2008)
Min, K. J., Yamamoto, R., Buch, S., Pankratz, M. & Tatar, M. Drosophila lifespan control by dietary restriction independent of insulin-like signaling. Aging Cell 7, 199–206 (2008)
Yuan, J. S., Reed, A., Chen, F. & Stewart, C. N., Jr Statistical analysis of real-time PCR data. BMC Bioinformatics 7, 85 (2006)
Brennecke, J. et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila . Cell 128, 1089–1103 (2007)
Haley, B. & Zamore, P. D. Kinetic analysis of the RNAi enzyme complex. Nature Struct. Mol. Biol. 11, 599–606 (2004)
Haley, B., Foys, B. & Levine, M. Vectors and parameters that enhance the efficacy of RNAi-mediated gene disruption in transgenic Drosophila . Proc. Natl Acad. Sci. USA 107, 11435–11440 (2010)
Acknowledgements
We are grateful to D. St Johnston, W. Chia, P. Macdonald, R. Carthew, E. Bach, D. Harrison, T. Volk, U. Heberlein, A. Spradling, J. Kadonaga, G. Hannon, T. Hays, N. Sokol, H. Siomi and P. Lasko for reagents and fly stocks, to C. Doe, R. Hans, G. Volohonsky, T. Juven-Gershon, A. Pasquinelli, R. Zhou and S. Aigner for guidance on methods used in this manuscript, to O. Tam for computational support, and to C. Koehler for technical assistance. This work was supported by the G. Harold and Leila Y. Mathers Charitable Foundation, the Ellison Medical Foundation, the Emerald Foundation, the American Federation for Aging Research, and the National Institutes of Health (to D.L.J.). B.C. is supported by a PhD fellowship from the Boehringer Ingelheim Fonds. E.L. is supported by the National Science Foundation.
Author information
Authors and Affiliations
Contributions
H.T., C.D. and D.L.J. designed experiments. H.T. and C.D. performed experiments. B.C. generated and analysed small RNA libraries. E.L. performed bioinformatic analysis to identify Imp-binding sequences. H.T., C.D., B.C. and D.L.J. evaluated the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-6. (PDF 16098 kb)
Supplementary Tables
This file contains Supplementary Table 1. (XLS 247 kb)
Rights and permissions
About this article
Cite this article
Toledano, H., D’Alterio, C., Czech, B. et al. The let-7–Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature 485, 605–610 (2012). https://doi.org/10.1038/nature11061
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11061
This article is cited by
-
Imp is expressed in INPs and newborn neurons where it regulates neuropil targeting in the central complex
Neural Development (2023)
-
Characterization of MicroRNA expression profiles in the ovarian tissue of goats during the sexual maturity period
Journal of Ovarian Research (2023)
-
Mesenchymal stem cells ameliorate cisplatin-induced acute kidney injury via let-7b-5p
Cell and Tissue Research (2023)
-
The Mechanism of Stem Cell Aging
Stem Cell Reviews and Reports (2022)
-
Molecular basis of reproductive senescence: insights from model organisms
Journal of Assisted Reproduction and Genetics (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.