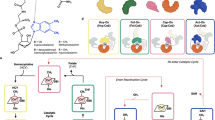
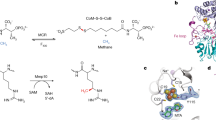
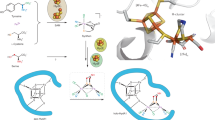
Abstract
Derivatives of vitamin B12 are used in methyl group transfer in biological processes as diverse as methionine synthesis in humans and CO2 fixation in acetogenic bacteria1,2,3. This seemingly straightforward reaction requires large, multimodular enzyme complexes that adopt multiple conformations to alternately activate, protect and perform catalysis on the reactive B12 cofactor. Crystal structures determined thus far have provided structural information for only fragments of these complexes4,5,6,7,8,9,10,11,12, inspiring speculation about the overall protein assembly and conformational movements inherent to activity. Here we present X-ray crystal structures of a complete 220 kDa complex that contains all enzymes responsible for B12-dependent methyl transfer, namely the corrinoid iron–sulphur protein and its methyltransferase from the model acetogen Moorella thermoacetica. These structures provide the first three-dimensional depiction of all protein modules required for the activation, protection and catalytic steps of B12-dependent methyl transfer. In addition, the structures capture B12 at multiple locations between its ‘resting’ and catalytic positions, allowing visualization of the dramatic protein rearrangements that enable methyl transfer and identification of the trajectory for B12 movement within the large enzyme scaffold. The structures are also presented alongside in crystallo spectroscopic data, which confirm enzymatic activity within crystals and demonstrate the largest known conformational movements of proteins in a crystalline state. Taken together, this work provides a model for the molecular juggling that accompanies turnover and helps explain why such an elaborate protein framework is required for such a simple, yet biologically essential reaction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Matthews, R. G. Cobalamin-dependent methyltransferases. Acc. Chem. Res. 34, 681–689 (2001)
Banerjee, R. B. & Ragsdale, S. W. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu. Rev. Biochem. 72, 209–247 (2003)
Matthews, R. G., Koutmos, M. & Datta, S. Cobalamin-dependent and cobamide-dependent methyltransferases. Curr. Opin. Struct. Biol. 18, 658–666 (2008)
Drennan, C. L., Huang, S., Drummond, J. T., Matthews, R. G. & Ludwig, M. L. How a protein binds B12: a 3.0 Å X-ray structure of B12-binding domains of methionine synthase. Science 266, 1669–1674 (1994)
Dixon, M. M., Huang, S., Matthews, R. G. & Ludwig, M. L. The structure of the C-terminal domain of methionine synthase: presenting S-adenosylmethionine for reductive methylation of B12. Structure 4, 1263–1275 (1996)
Doukov, T., Seravalli, J., Stezowski, J. J. & Ragsdale, S. W. Crystal structure of a methyltetrahydrofolate- and corrinoid-dependent methyltransferase. Structure 8, 817–830 (2000)
Bandarian, V. et al. Domain alternation switches B12-dependent methionine synthase to the active conformation. Nature Struct. Biol. 9, 53–56 (2002)
Evans, J. C. et al. Structures of the N-terminal module imply large domain motions during catalysis by methionine synthase. Proc. Natl Acad. Sci. USA 101, 3729–3736 (2004)
Svetlitchnaia, T., Svetlitchnyi, V., Meyer, O. & Dobbek, H. Structural insights into methyltransfer reactions of a corrinoid iron-sulfur protein involved in acetyl-CoA synthesis. Proc. Natl Acad. Sci. USA 103, 14331–14336 (2006)
Doukov, T. I., Hemmi, H., Drennan, C. L. & Ragsdale, S. W. Structural and kinetic evidence for an extended hydrogen-bonding network in catalysis of methyl group transfer: role of an active site asparagine residue in activation of methyl transfer by methyltransferases. J. Biol. Chem. 282, 6609–6618 (2007)
Datta, S., Koutmos, M., Pattridge, K. A., Ludwig, M. L. & Matthews, R. G. A. Disulfide-stabilized conformer of methionine synthase reveals an unexpected role for the histidine ligand of the cobalamin cofactor. Proc. Natl Acad. Sci. USA 105, 4115–4120 (2008)
Koutmos, M., Datta, S., Pattridge, K. A., Smith, J. L. & Matthews, R. G. Insights into the reactivation of cobalamin-dependent methionine synthase. Proc. Natl Acad. Sci. USA 106, 18527–18532 (2009)
Banerjee, R. B. & Matthews, R. G. Cobalamin-dependent methionine synthase. FASEB J. 4, 1450–1459 (1990)
Ragsdale, S. W. & Pierce, E. Acetogenesis and the Wood–Ljungdahl pathway of CO2 fixation. Biochim. Biophys. Acta 1784, 1873–1898 (2008)
Schrauzer, G. N. & Deutsch, E. Reactions of cobalt(I) supernucleophiles. The alkylations of vitamin B12s, cobaloximes(I), and related compounds. J. Am. Chem. Soc. 91, 3341–3350 (1969)
Harder, S. R., Lu, W.-P., Feinberg, B. A. & Ragsdale, S. W. Spectroelectrochemical studies of the corrinoid/iron-sulfur protein involved in acetyl coenzyme a synthesis by Clostridium thermoaceticum. Biochemistry 28, 9080–9087 (1989)
Banerjee, R. B., Harder, S. R., Ragsdale, S. W. & Matthews, R. G. Mechanism of reductive activation of cobalamin-dependent methionine synthase: an electron paramagnetic resonance spectroelectrochemical study. Biochemistry 29, 1129–1135 (1990)
Menon, S. & Ragsdale, S. W. Role of the [4Fe-4S] cluster in reductive activation of the cobalt center of the corrinoid iron–sulfur protein from Clostridium thermoaceticum during acetate biosynthesis. Biochemistry 37, 5689–5698 (1998)
Menon, S. & Ragsdale, S. W. The role of an iron–sulfur cluster in an enzymatic methylation reaction. J. Biol. Chem. 274, 11513–11518 (1999)
Zydowsky, T. M. et al. Stereochemical analysis of the methyl transfer catalyzed by cobalamin-dependent methionine synthase from Escherichia coli B. J. Am. Chem. Soc. 108, 3152–3153 (1986)
Ragsdale, S. W., Lindahl, P. A. & Münck, E. Mössbauer, EPR, and optical studies of the corrinoid/iron–sulfur protein involved in the synthesis of acetyl coenzyme A by Clostridium thermoaceticum. J. Biol. Chem. 262, 14289–14297 (1987)
Zhao, S., Roberts, D. L. & Ragsdale, S. W. Mechanistic studies of the methyltransferase from Clostridium thermoaceticum: origin of the pH dependence of the methyl group transfer from methyltetrahydrofolate to the corrinoid/iron–sulfur protein. Biochemistry 34, 15075–15083 (1995)
Seravalli, J., Zhao, S. & Ragsdale, S. W. Mechanism of transfer of the methyl group from (6S)-methytetrahydrofolate to the corrinoid/iron–sulfur protein catalyzed by the methyltransferase from Clostridium thermoaceticum: a key step in the Wood–Ljungdahl pathway of acetyl-CoA synthesis. Biochemistry 38, 5728–5735 (1999)
Jarrett, J. T. et al. Mutations in the B12-binding region of methionine synthase: how the protein controls methylcobalamin reactivity. Biochemistry 35, 2464–2475 (1996)
Bandarian, V., Ludwig, M. L. & Matthews, R. G. Factors modulating conformational equilibria in large modular proteins: a case study with cobalamin-dependent methionine synthase. Proc. Natl Acad. Sci. USA 100, 8156–8163 (2003)
Wirt, M. D. et al. Structural and electronic factors in heterolytic cleavage: formation of the Co(I) intermediate in the corrinoid/iron–sulfur protein from Clostridium thermoaceticum. Biochemistry 34, 5269–5273 (1995)
Stich, T. A. et al. Spectroscopic studies of the corrinoid/iron–sulfur protein from Moorella thermoacetica. J. Am. Chem. Soc. 128, 5010–5020 (2006)
Elliott, J. I. & Brewer, J. M. The inactivation of yeast enolase by 2,3-butanedione. Arch. Biochem. Biophys. 190, 351–357 (1978)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Brünger, A. T. et al. Crystallography & NMR System: a new software suite for marcromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Adams, P. D. et al. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Laskowski, R. A., MacArthur, M. W. & Thornton, J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993)
Lu, W.-P., Schiau, I., Cunningham, J. R. & Ragsdale, S. W. Sequence and expression of the gene encoding the corrinoid/iron–sulfur protein from Clostridium thermoaceticum and reconsitution of the recombinant protein to full activity. J. Biol. Chem. 268, 5605–5614 (1993)
Ljungdahl, L. G., LeGall, J. & Lee, J.-P. Isolation of a protein containing tightly bound 5-methoxybenzimidazolylcobamide (factor IIIm) from Clostridium thermoaceticum. Biochemistry 12, 1802–1808 (1973)
Zehnder, A. J. B. & Wuhrmann, K. Titanium(III) citrate as a nontoxic oxidation-reduction buffering system for the culture of obligate anaerobes. Science 194, 1165–1166 (1976)
Royant, A. et al. Advances in spectroscopic methods for biological crystals. 1. fluorescence lifetime measurements. J. Appl. Cryst. 40, 1105–1112 (2007)
Barstow, B., Ando, N., Kim, C. U. & Gruner, S. M. Alteration of citrine structure by hydrostatic pressure explains the accompanying spectral shift. Proc. Natl Acad. Sci. USA 105, 13362–13366 (2008)
Acknowledgements
We thank J. E. Darty for his assistance with the purification of CFeSP. This work was supported by National Institutes of Health grants GM69857 (to C.L.D.) and GM39451 (to S.W.R.) and the MIT Energy Initiative (to C.L.D.). C.L.D. is a Howard Hughes Medical Institute Investigator. This work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the US Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under Contract No. DE-AC02-05CH11231.
Author information
Authors and Affiliations
Contributions
Y.K. performed crystallization and data collection, processing and refinement that gave the folate-free and folate-bound CFeSP/MeTr structures. N.A. built the microspectrophotometer and performed in crystallo spectroscopic experiments with the aid of Y.K., who performed the parallel solution spectroscopic experiments. T.I.D. determined initial crystallization conditions and performed initial data collection, and L.C.B. processed and refined these data. G.B. and J.S. expressed and purified protein samples, and S.W.R. and C.L.D. were involved in study design. Y.K. and C.L.D. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-2, containing X-ray data collection and refinement statistics and B-factor information, Supplementary Figures 1-21, a Supplementary Discussion and additional references. (PDF 21726 kb)
Rights and permissions
About this article
Cite this article
Kung, Y., Ando, N., Doukov, T. et al. Visualizing molecular juggling within a B12-dependent methyltransferase complex. Nature 484, 265–269 (2012). https://doi.org/10.1038/nature10916
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10916
This article is cited by
-
Insights into pyrrolysine function from structures of a trimethylamine methyltransferase and its corrinoid protein complex
Communications Biology (2023)
-
Structure of full-length cobalamin-dependent methionine synthase and cofactor loading captured in crystallo
Nature Communications (2023)
-
Structure determination of the HgcAB complex using metagenome sequence data: insights into microbial mercury methylation
Communications Biology (2020)
-
Genome-scale analysis of syngas fermenting acetogenic bacteria reveals the translational regulation for its autotrophic growth
BMC Genomics (2018)
-
Orchestrated Domain Movement in Catalysis by Cytochrome P450 Reductase
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.