Abstract
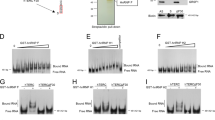
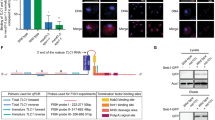
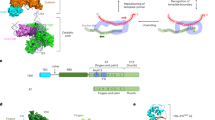
In most eukaryotes, the progressive loss of chromosome-terminal DNA sequences is counteracted by the enzyme telomerase, a reverse transcriptase that uses part of an RNA subunit as template to synthesize telomeric repeats. Many cancer cells express high telomerase activity, and mutations in telomerase subunits are associated with degenerative syndromes including dyskeratosis congenita and aplastic anaemia. The therapeutic value of altering telomerase activity thus provides ample impetus to study the biogenesis and regulation of this enzyme in human cells and model systems. We have previously identified a precursor of the fission yeast telomerase RNA subunit (TER1)1 and demonstrated that the mature 3′-end is generated by the spliceosome in a single cleavage reaction akin to the first step of splicing2. Directly upstream and partly overlapping with the spliceosomal cleavage site is a putative binding site for Sm proteins. Sm and like-Sm (LSm) proteins belong to an ancient family of RNA-binding proteins represented in all three domains of life3. Members of this family form ring complexes on specific sets of target RNAs and have critical roles in their biogenesis, function and turnover. Here we demonstrate that the canonical Sm ring and the Lsm2–8 complex sequentially associate with fission yeast TER1. The Sm ring binds to the TER1 precursor, stimulates spliceosomal cleavage and promotes the hypermethylation of the 5′-cap by Tgs1. Sm proteins are then replaced by the Lsm2–8 complex, which promotes the association with the catalytic subunit and protects the mature 3′-end of TER1 from exonucleolytic degradation. Our findings define the sequence of events that occur during telomerase biogenesis and characterize roles for Sm and Lsm complexes as well as for the methylase Tgs1.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Leonardi, J., Box, J. A., Bunch, J. T. & Baumann, P. TER1, the RNA subunit of fission yeast telomerase. Nature Struct. Mol. Biol. 15, 26–33 (2008)
Box, J. A., Bunch, J. T., Tang, W. & Baumann, P. Spliceosomal cleavage generates the 3′ end of telomerase RNA. Nature 456, 910–914 (2008)
Wilusz, C. J. & Wilusz, J. Eukaryotic Lsm proteins: lessons from bacteria. Nature Struct. Mol. Biol. 12, 1031–1036 (2005)
Raker, V. A., Plessel, G. & Lührmann, R. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 15, 2256–2269 (1996)
Patel, S. B. & Bellini, M. The assembly of a spliceosomal small nuclear ribonucleoprotein particle. Nucleic Acids Res. 36, 6482–6493 (2008)
Terns, M. P. & Terns, R. M. Macromolecular complexes: SMN — the master assembler. Curr. Biol. 11, R862–R864 (2001)
Tharun, S. et al. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404, 515–518 (2000)
Bouveret, E., Rigaut, G., Shevchenko, A., Wilm, M. & Seraphin, B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 19, 1661–1671 (2000)
Achsel, T. et al. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 18, 5789–5802 (1999)
Mayes, A. E., Verdone, L., Legrain, P. & Beggs, J. D. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 18, 4321–4331 (1999)
Kufel, J., Allmang, C., Verdone, L., Beggs, J. D. & Tollervey, D. Lsm proteins are required for normal processing of pre-tRNAs and their efficient association with La-homologous protein Lhp1p. Mol. Cell. Biol. 22, 5248–5256 (2002)
Kufel, J., Allmang, C., Petfalski, E., Beggs, J. & Tollervey, D. Lsm proteins are required for normal processing and stability of ribosomal RNAs. J. Biol. Chem. 278, 2147–2156 (2003)
Dandjinou, A. T. et al. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr. Biol. 14, 1148–1158 (2004)
Gunisova, S. et al. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA 15, 546–559 (2009)
Seto, A. G., Zaug, A. J., Sobel, S. G., Wolin, S. L. & Cech, T. R. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401, 177–180 (1999)
Webb, C. J. & Zakian, V. A. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nature Struct. Mol. Biol. 15, 34–42 (2008)
Yong, J., Kasim, M., Bachorik, J. L., Wan, L. & Dreyfuss, G. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol. Cell 38, 551–562 (2010)
Jady, B. E., Bertrand, E. & Kiss, T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell Biol. 164, 647–652 (2004)
Mattaj, I. W. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell 46, 905–911 (1986)
Plessel, G., Fischer, U. & Lührmann, R. m3G cap hypermethylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP cytoplasmic protein that requires a binding site on the Sm core domain. Mol. Cell. Biol. 14, 4160–4172 (1994)
Mouaikel, J., Verheggen, C., Bertrand, E., Tazi, J. & Bordonne, R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell 9, 891–901 (2002)
Hausmann, S., Ramirez, A., Schneider, S., Schwer, B. & Shuman, S. Biochemical and genetic analysis of RNA cap guanine-N2 methyltransferases from Giardia lamblia and Schizosaccharomyces pombe. Nucleic Acids Res. 35, 1411–1420 (2007)
Franke, J., Gehlen, J. & Ehrenhofer-Murray, A. E. Hypermethylation of yeast telomerase RNA by the snRNA and snoRNA methyltransferase Tgs1. J. Cell Sci. 121, 3553–3560 (2008)
Harrington, L. Making the most of a little: dosage effects in eukaryotic telomere length maintenance. Chromosome Res. 13, 493–504 (2005)
Mozdy, A. D. & Cech, T. R. Low abundance of telomerase in yeast: implications for telomerase haploinsufficiency. RNA 12, 1721–1737 (2006)
Berman, A. J., Gooding, A. R. & Cech, T. R. Tetrahymena telomerase protein p65 induces conformational changes throughout telomerase RNA (TER) and rescues telomerase reverse transcriptase and TER assembly mutants. Mol. Cell. Biol. 30, 4965–4976 (2010)
O’Connor, C. M. & Collins, K. A novel RNA binding domain in tetrahymena telomerase p65 initiates hierarchical assembly of telomerase holoenzyme. Mol. Cell. Biol. 26, 2029–2036 (2006)
Stone, M. D. et al. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature 446, 458–461 (2007)
Fu, D. & Collins, K. Human telomerase and Cajal body ribonucleoproteins share a unique specificity of Sm protein association. Genes Dev. 20, 531–536 (2006)
Fu, D. & Collins, K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol. Cell 28, 773–785 (2007)
Bahler, J. et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 (1998)
Bunch, J. T., Bae, N. S., Leonardi, J. & Baumann, P. Distinct requirements for Pot1 in limiting telomere length and maintaining chromosome stability. Mol. Cell. Biol. 25, 5567–5578 (2005)
Haering, C. H., Nakamura, T. M., Baumann, P. & Cech, T. R. Analysis of telomerase catalytic subunit mutants in vivo and in vitro in Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA 97, 6367–6372 (2000)
Acknowledgements
We thank S. Shuman for the tgs1Δ strain, J. A. Box, J. T. Bunch, S. Hartnett and R. M. Helston for technical assistance, the Molecular Biology Core Facility for site-directed mutagenesis and sequencing, D. P. Baumann and R. M. Helston for proofreading the manuscript, and all members of the Baumann laboratory for discussions. This work was funded in part by the Stowers Institute for Medical Research. R.K. is supported by an award from the American Heart Association, and P.B. is an Early Career Scientist with the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
P.B. and W.T. conceived the study and designed the experiments; W.T. performed most of the experiments with some assistance from those acknowledged and P.B.; R.K. contributed to the characterization of Sm mutants and analysed telomere length of Myc-tagged strains. M.B. wrote the script for sequence data analysis and provided advice; W.T., R.K. and P.B. analysed the data, and W.T. and P.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-6 and Supplementary Table 1. (PDF 2662 kb)
Rights and permissions
About this article
Cite this article
Tang, W., Kannan, R., Blanchette, M. et al. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature 484, 260–264 (2012). https://doi.org/10.1038/nature10924
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10924
This article is cited by
-
The mechanism of LSM2 in the progression of live hepatocellular carcinoma was analyzed based on bioinformatics
Medical Oncology (2023)
-
The methyl phosphate capping enzyme Bmc1/Bin3 is a stable component of the fission yeast telomerase holoenzyme
Nature Communications (2022)
-
Maturation and shuttling of the yeast telomerase RNP: assembling something new using recycled parts
Current Genetics (2022)
-
Regulation of human telomerase in homeostasis and disease
Nature Reviews Molecular Cell Biology (2020)
-
LARP7 family proteins have conserved function in telomerase assembly
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.