Abstract
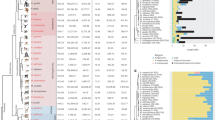
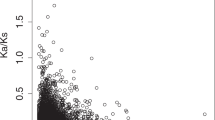
The human X and Y chromosomes evolved from an ordinary pair of autosomes during the past 200–300 million years1,2,3. The human MSY (male-specific region of Y chromosome) retains only three percent of the ancestral autosomes’ genes owing to genetic decay4,5. This evolutionary decay was driven by a series of five ‘stratification’ events. Each event suppressed X–Y crossing over within a chromosome segment or ‘stratum’, incorporated that segment into the MSY and subjected its genes to the erosive forces that attend the absence of crossing over2,6. The last of these events occurred 30 million years ago, 5 million years before the human and Old World monkey lineages diverged. Although speculation abounds regarding ongoing decay and looming extinction of the human Y chromosome7,8,9,10, remarkably little is known about how many MSY genes were lost in the human lineage in the 25 million years that have followed its separation from the Old World monkey lineage. To investigate this question, we sequenced the MSY of the rhesus macaque, an Old World monkey, and compared it to the human MSY. We discovered that during the last 25 million years MSY gene loss in the human lineage was limited to the youngest stratum (stratum 5), which comprises three percent of the human MSY. In the older strata, which collectively comprise the bulk of the human MSY, gene loss evidently ceased more than 25 million years ago. Likewise, the rhesus MSY has not lost any older genes (from strata 1–4) during the past 25 million years, despite its major structural differences to the human MSY. The rhesus MSY is simpler, with few amplified gene families or palindromes that might enable intrachromosomal recombination and repair. We present an empirical reconstruction of human MSY evolution in which each stratum transitioned from rapid, exponential loss of ancestral genes to strict conservation through purifying selection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Primary accessions
GenBank/EMBL/DDBJ
Data deposits
cDNA sequences of rhesus Y genes have been deposited in GenBank (http://www.ncbi.nlm.nih.gov) under accession numbers FJ527009–FJ527028 and FJ648737–FJ648739. 454 testis cDNA sequences have been deposited in GenBank under accession number SRA039857.
References
Charlesworth, B. The evolution of sex chromosomes. Science 251, 1030–1033 (1991)
Lahn, B. T. & Page, D. C. Four evolutionary strata on the human X chromosome. Science 286, 964–967 (1999)
Ross, M. T. et al. The DNA sequence of the human X chromosome. Nature 434, 325–337 (2005)
Skaletsky, H. et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 (2003)
Bellott, D. W. et al. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 466, 612–616 (2010)
Charlesworth, B. & Charlesworth, D. The degeneration of Y chromosomes. Phil. Trans. R. Soc. Lond. B 355, 1563–1572 (2000)
Aitken, R. J. & Marshall Graves, J. A. The future of sex. Nature 415, 963 (2002)
Sykes, B. Adam's Curse (Norton, W. W. & Company, 2004)
Graves, J. A., Koina, E. & Sankovic, N. How the gene content of human sex chromosomes evolved. Curr. Opin. Genet. Dev. 16, 219–224 (2006)
Marshall Graves, J. A. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu. Rev. Genet. 42, 565–586 (2008)
Kuroda-Kawaguchi, T. et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nature Genet. 29, 279–286 (2001)
Hughes, J. F. et al. Conservation of Y-linked genes during human evolution revealed by comparative sequencing in chimpanzee. Nature 437, 100–103 (2005)
Hughes, J. F. et al. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 463, 536–539 (2010)
Gibbs, R. A. et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science 316, 222–234 (2007)
Miyata, T., Hayashida, H., Kuma, K., Mitsuyasu, K. & Yasunaga, T. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harb. Symp. Quant. Biol. 52, 863–867 (1987)
Bohossian, H. B., Skaletsky, H. & Page, D. C. Unexpectedly similar rates of nucleotide substitution found in male and female hominids. Nature 406, 622–625 (2000)
Rozen, S. et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423, 873–876 (2003)
Lahn, B. T. & Page, D. C. Functional coherence of the human Y chromosome. Science 278, 675–680 (1997)
Lahn, B. T. & Page, D. A human sex-chromosomal gene family expressed in male germ cells and encoding variably charged proteins. Hum. Mol. Genet. 9, 311–319 (2000)
Rozen, S., Marszalek, J. D., Alagappan, R. K., Skaletsky, H. & Page, D. C. Remarkably little variation in proteins encoded by the Y chromosome’s single-copy genes, implying effective purifying selection. Am. J. Hum. Genet. 85, 923–928 (2009)
Dixson, A. F. Primate Sexuality: Comparative Studies of the Prosimians, Monkeys, Apes and Human Beings (Univ. Chicago Press, 1998)
Rice, W. R. Genetic hitchhiking and the evolution of reduced genetic activity of the Y sex chromosome. Genetics 116, 161–167 (1987)
Schultz, A. H. The relative weight of the testes in primates. Anat. Rec. 72, 387–394 (1938)
Harcourt, A. H., Harvey, P. H., Larson, S. G. & Short, R. V. Testis weight, body weight and breeding system in primates. Nature 293, 55–57 (1981)
Graves, J. A. Sex chromosome specialization and degeneration in mammals. Cell 124, 901–914 (2006)
Bachtrog, D. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics 179, 1513–1525 (2008)
Saxena, R. et al. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nature Genet. 14, 292–299 (1996)
Karere, G. M., Froenicke, L., Millon, L., Womack, J. E. & Lyons, L. A. A high-resolution radiation hybrid map of rhesus macaque chromosome 5 identifies rearrangements in the genome assembly. Genomics 92, 210–218 (2008)
Slonim, D., Kruglyak, L., Stein, L. & Lander, E. Building human genome maps with radiation hybrids. J. Comput. Biol. 4, 487–504 (1997)
Shimmin, L. C., Chang, B. H. & Li, W. H. Male-driven evolution of DNA sequences. Nature 362, 745–747 (1993)
Acknowledgements
We thank V. Frazzoni, G. Rogers and S. Zaghlul for technical assistance; L. Lyons, W. Murphy and J. Womack for radiation hybrid panels; W. Johnson, E. Curran, S. O’Neil and A. Kaur for tissue samples; R. Harris for analyses of rhesus–human genome alignments; T. DiCesare for assistance with figures; and D. Bellott, M. Carmell, R. Desgraz, Y. Hu, B. Lesch, J. Mueller, K. Romer and S. Soh for comments on the manuscript. This work was supported by the National Institutes of Health, the Howard Hughes Medical Institute and the Charles A. King Trust.
Author information
Authors and Affiliations
Contributions
J.F.H., H.Sk., W.C.W., S.R., R.A.G., R.K.W. and D.C.P. planned the project. J.F.H., H.Sk., L.G.B., T.J.C. and N.K. performed BAC mapping, radiation hybrid mapping and real-time polymerase chain reaction analyses. T.G., R.S.F., S.D., Y.D., C.J.B., C.K., Q.W., H.Sh., M.H., D.V., L.V.N., A.C., L.C., J.V., H.K. and D.M.M. were responsible for BAC sequencing. J.F.H. and H.Sk. performed comparative sequence analyses. T.P. performed FISH analyses. J.F.H. and D.C.P. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Notes 1-3, Supplementary Figures 1-16 with legends and Supplementary Tables 1-8. (PDF 16750 kb)
Supplementary Data 1
This is a FASTA file of rhesus MSY sequence assembly. (TXT 11806 kb)
Supplementary Data 2
This file contains radiation hybrid mapping data vectors. (TXT 1 kb)
Supplementary Data 3
This file shows the alignment of rhesus and human MSY sequences in FASTA format. (TXT 9279 kb)
Rights and permissions
About this article
Cite this article
Hughes, J., Skaletsky, H., Brown, L. et al. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature 483, 82–86 (2012). https://doi.org/10.1038/nature10843
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10843
This article is cited by
-
Assessing the recovery of Y chromosome microsatellites with population genomic data using Papio and Theropithecus genomes
Scientific Reports (2023)
-
Eighty million years of rapid evolution of the primate Y chromosome
Nature Ecology & Evolution (2023)
-
A gene deriving from the ancestral sex chromosomes was lost from the X and retained on the Y chromosome in eutherian mammals
BMC Biology (2022)
-
Sex differences in the meiotic behavior of an XX sex chromosome pair in males and females of the mole vole Ellobius tancrei: turning an X into a Y chromosome?
Chromosoma (2021)
-
A Hu sheep genome with the first ovine Y chromosome reveal introgression history after sheep domestication
Science China Life Sciences (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.