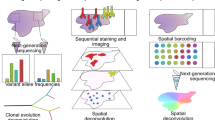
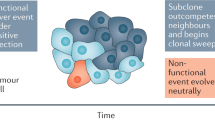
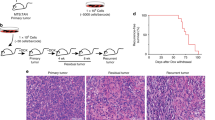
Abstract
Cancers evolve by a reiterative process of clonal expansion, genetic diversification and clonal selection within the adaptive landscapes of tissue ecosystems. The dynamics are complex, with highly variable patterns of genetic diversity and resulting clonal architecture. Therapeutic intervention may destroy cancer clones and erode their habitats, but it can also inadvertently provide a potent selective pressure for the expansion of resistant variants. The inherently Darwinian character of cancer is the primary reason for this therapeutic failure, but it may also hold the key to more effective control.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Jemal, A. et al. Cancer statistics, 2008. CA Cancer J. Clin. 58, 71–96 (2008).
Stratton, M. R. Exploring the genomes of cancer cells: progress and promise. Science 331, 1553–1558 (2011).
Nowell, P. C. The clonal evolution of tumor cell populations. Science 194, 23–28 (1976). The foundation paper that established the evolutionary theory of cancer.
Merlo, L. M., Pepper, J. W., Reid, B. J. & Maley, C. C. Cancer as an evolutionary and ecological process. Nature Rev. Cancer 6, 924–935 (2006).
Pepper, J., Scott Findlay, C., Kassen, R., Spencer, S. & Maley, C. Cancer research meets evolutionary biology. Evol. Appl. 2, 62–70 (2009).
Greaves, M. Cancer: The Evolutionary Legacy (Oxford Univ. Press, 2000).
Sakr, W. A., Haas, G. P., Cassin, B. F., Pontes, J. E. & Crissman, J. D. The frequency of carcinoma and intraepithelial neoplasia of the prostate in young male patients. J. Urol. 150, 379–385 (1993).
Mori, H. et al. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc. Natl Acad. Sci. USA 99, 8242–8247 (2002).
Reid, B. J., Li, X., Galipeau, P. C. & Vaughan, T. L. Barrett's oesophagus and oesophageal adenocarcinoma: time for a new synthesis. Nature Rev. Cancer 10, 87–101 (2010).
Klein, C. A. Parallel progression of primary tumours and metastases. Nature Rev. Cancer 9, 302–312 (2009).
Malaise, E. P., Chavaudra, N. & Tubiana, M. The relationship between growth rate, labelling index and histological type of human solid tumours. Eur. J. Cancer 9, 305–312 (1973).
Tsai, A. G. et al. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell 135, 1130–1142 (2008).
Bardelli, A. et al. Carcinogen-specific induction of genetic instability. Proc. Natl Acad. Sci. USA 98, 5770–5775 (2001).
Cahill, D. P., Kinzler, K. W., Vogelstein, B. & Lengauer, C. Genetic instability and Darwinian selection in tumors. Trends Cell Biol. 9, M57–M60 (1999).
Barcellos-Hoff, M. H., Park, C. & Wright, E. G. Radiation and the microenvironment – tumorigenesis and therapy. Nature Rev. Cancer 5, 867–875 (2005).
Maley, C. C. et al. Selectively advantageous mutations and hitchhikers in neoplasms: p16 lesions are selected in Barrett's esophagus. Cancer Res. 64, 3414–3427 (2004).
Tao, Y. et al. Rapid growth of a hepatocellular carcinoma and the driving mutations revealed by cell-population genetic analysis of whole-genome data. Proc. Natl Acad. Sci. USA 108, 12042–12047 (2011).
Bignell, G. R. et al. Signatures of mutation and selection in the cancer genome. Nature 463, 893–898 (2010).
Youn, A. & Simon, R. Identifying cancer driver genes in tumor genome sequencing studies. Bioinformatics 27, 175–181 (2011).
Greenman, C., Wooster, R., Futreal, P. A., Stratton, M. R. & Easton, D. F. Statistical analysis of pathogenicity of somatic mutations in cancer. Genetics 173, 2187–2198 (2006).
Bozic, I. et al. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl Acad. Sci. USA 107, 18545–18550 (2010).
Schwartz, M., Zlotorynski, E. & Kerem, B. The molecular basis of common and rare fragile sites. Cancer Lett. 232, 13–26 (2006).
Loeb, L. A. Human cancers express mutator phenotypes: origin, consequences and targeting. Nature Rev. Cancer 11, 450–457 (2011).
Weisenberger, D. J. et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nature Genet 38, 787–793 (2006).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). This paper consolidates the common phenotypes that evolve in neoplastic cells of all types.
Siegmund, K. D., Marjoram, P., Woo, Y. J., Tavare, S. & Shibata, D. Inferring clonal expansion and cancer stem cell dynamics from DNA methylation patterns in colorectal cancers. Proc. Natl Acad. Sci. USA 106, 4828–4833 (2009).
Varley, K. E., Mutch, D. G., Edmonston, T. B., Goodfellow, P. J. & Mitra, R. D. Intra-tumor heterogeneity of MLH1 promoter methylation revealed by deep single molecule bisulfite sequencing. Nucleic Acids Res. 37, 4603–4612 (2009).
Aktipis, C. A., Kwan, V. S. Y., Johnson, K. A., Neuberg, S. L. & Maley, C. C. Overlooking evolution: a systematic analysis of cancer relapse and therapeutic resistance research. PLoS ONE 6, e261000 10.1371/journal.pone.0026100 (2011).
Beerenwinkel, N. et al. Genetic progression and the waiting time to cancer. PLoS Comput. Biol. 3, e225 (2007).
de Visser, J. A. & Rozen, D. E. Clonal interference and the periodic selection of new beneficial mutations in Escherichia coli . Genetics 172, 2093–2100 (2006).
Leedham, S. J. et al. Individual crypt genetic heterogeneity and the origin of metaplastic glandular epithelium in human Barrett's oesophagus. Gut 57, 1041–1048 (2008).
Navin, N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011). Single-cell sequencing revealed the clonal structure of two breast cancers.
Anderson, K. et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469, 356–361 (2011). Single-cell genetic analyses and xenografts revealed the clonal architecture within acute lymphoblastic leukaemia stem-cell populations and demonstrated repeated independent acquisition of copy number changes within the same neoplasm.
Tsao, J. L. et al. Colorectal adenoma and cancer divergence. Evidence of multilineage progression. Am. J. Pathol. 154, 1815–1824 (1999).
Maley, C. C. et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nature Genet. 38, 468–473 (2006).
Sidransky, D. et al. Clonal expansion of p53 mutant cells is associated with brain tumour progression. Nature 355, 846–847 (1992).
Yachida, S. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010).
Gould, S. J. & Eldredge, N. Punctuated equilibrium comes of age. Nature 366, 223–227 (1993).
Stephens, P. J. et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40 (2011).
Campbell, P. J. et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc. Natl Acad. Sci. USA 105, 13081–13086 (2008). Deep sequencing revealed rare (frequency <0.001) intermediate genotypes between the common clones in leukaemias (using immunoglobulin rearrangements as surrogate mutations).
Aguirre-Ghiso, J. A. Models, mechanisms and clinical evidence for cancer dormancy. Nature Rev. Cancer 7, 834–846 (2007).
Isoda, T. et al. Immunologically silent cancer clone transmission from mother to offspring. Proc. Natl Acad. Sci. USA 106, 17882–17885 (2009).
Welsh, J. S. Contagious cancer. Oncologist 16, 1–4 (2011).
Gatenby, R. A. & Gillies, R. J. A microenvironmental model of carcinogenesis. Nature Rev. Cancer 8, 56–61 (2008).
Bierie, B. & Moses, H. L. Tumour microenvironment: TGFβ: the molecular Jekyll and Hyde of cancer. Nature Rev. Cancer 6, 506–520 (2006).
Lathia, J. D., Heddleston, J. M., Venere, M. & Rich, J. N. Deadly teamwork: neural cancer stem cells and the tumor microenvironment. Cell Stem Cell 8, 482–485 (2011).
Cairns, J. Mutation selection and the natural history of cancer. Nature 255, 197–200 (1975). This paper identified natural selection as a driving force in carcinogenesis and identified tissue architecture as a cancer suppressor, and posited an immortal strand of DNA in tissue stem cells.
Anderson, A. R., Weaver, A. M., Cummings, P. T. & Quaranta, V. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell 127, 905–915 (2006).
Chen, J., Sprouffske, K., Huang, Q. & Maley, C. C. Solving the puzzle of metastasis: the evolution of cell migration in neoplasms. PLoS ONE 6, e17933 (2011).
Mazzone, M. et al. Heterozygous deficiency of PHD2 restores tumor oxygenation and inhibits metastasis via endothelial normalization. Cell 136, 839–851 (2009).
Gilbert, L. A. & Hemann, M. T. DNA damage-mediated induction of a chemoresistant niche. Cell 143, 355–366 (2010).
Jones, S. et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl Acad. Sci. USA 105, 4283–4288 (2008).
Ding, L. et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005 (2010).
Sprouffske, K., Pepper, J. W. & Maley, C. C. Accurate reconstruction of the temporal order of mutations in neoplastic progression. Cancer Prev. Res. 4, 1135–1144 (2011).
Greaves, M. F., Maia, A. T., Wiemels, J. L. & Ford, A. M. Leukemia in twins: lessons in natural history. Blood 102, 2321–2333 (2003).
Bateman, C. M. et al. Acquisition of genome-wide copy number alterations in monozygotic twins with acute lymphoblastic leukemia. Blood 115, 3553–3558 (2010).
Oosterhuis, J. W. & Looijenga, L. H. Testicular germ-cell tumours in a broader perspective. Nature Rev. Cancer 5, 210–222 (2005).
Grant, P. R. & Grant, B. R. How and Why Species Multiply (Princeton Univ. Press, 2008).
Durinck, S. et al. Temporal dissection of tumorigenesis in primary cancers. Cancer Discov. 1, 137–143 (2011).
Gonzalez-Garcia, I., Sole, R. V. & Costa, J. Metapopulation dynamics and spatial heterogeneity in cancer. Proc. Natl Acad. Sci. USA 99, 13085–13089 (2002).
Clark, J. et al. Complex patterns of ETS gene alteration arise during cancer development in the human prostate. Oncogene 27, 1993–2003 (2008).
Navin, N. et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 20, 68–80 (2010).
Allred, D. C. et al. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin. Cancer Res. 14, 370–378 (2008).
Park, S. Y., Gonen, M., Kim, H. J., Michor, F. & Polyak, K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J. Clin. Invest. 120, 636–644 (2010).
Merlo, L. M. et al. A comprehensive survey of clonal diversity measures in Barrett's esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prev. Res. 3, 1388–1397 (2010).
Dick, J. E. Stem cell concepts renew cancer research. Blood 112, 4793–4807 (2008).
Reya, T., Morrison, S. J., Clarke, M. F. & Weissman, I. L. Stem cells, cancer, and cancer stem cells. Nature 414, 105–111 (2001).
Greaves, M. Cancer stem cells renew their impact. Nature Med. 17, 1046–1048 (2011).
Rosen, J. M. & Jordan, C. T. The increasing complexity of the cancer stem cell paradigm. Science 324, 1670–1673 (2009).
Greaves, M. Cancer stem cells: back to Darwin? Semin. Cancer Biol. 20, 65–70 (2010).
Gupta, P. B. et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 138, 645–659 (2009).
Jamieson, C. H. et al. Granulocyte–macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N. Engl. J. Med. 351, 657–667 (2004).
Akala, O. O. et al. Long-term haematopoietic reconstitution by Trp53−/−p164−/−p19Arf−/− multipotent progenitors. Nature 453, 228–232 (2008).
Krivtsov, A. V. et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL–AF9. Nature 442, 818–822 (2006).
Olivier, M. & Taniere, P. Somatic mutations in cancer prognosis and prediction: lessons from TP53 and EGFR genes. Curr. Opin. Oncol. 23, 88–92 (2011).
Mizuno, H., Spike, B. T., Wahl, G. M. & Levine, A. J. Inactivation of p53 in breast cancers correlates with stem cell transcriptional signatures. Proc. Natl Acad. Sci. USA 107, 22745–22750 (2010).
Cicalese, A. et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 138, 1083–1095 (2009).
Quintana, E. et al. Efficient tumour formation by single human melanoma cells. Nature 456, 593–598 (2008). New xenograft methods revealed that cancer stem cells are common cell types in melanoma.
Pece, S. et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell 140, 62–73 (2010).
Notta, F. et al. Evolution of human BCR–ABL1 lymphoblastic leukaemia-initiating cells. Nature 469, 362–367 (2011).
Clappier, E. et al. Clonal selection in xenografted human T cell acute lymphoblastic leukemia recapitulates gain of malignancy at relapse. J. Exp. Med. 208, 653–661 (2011).
Frank, N. Y., Schatton, T. & Frank, M. H. The therapeutic promise of the cancer stem cell concept. J. Clin. Invest. 120, 41–50 (2010).
Ishikawa, F. et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nature Biotechnol. 25, 1315–1321 (2007).
Marusyk, A. & Polyak, K. Tumor heterogeneity: causes and consequences. Biochim. Biophys. Acta 1805, 105–117 (2010).
Piccirillo, S. G. M. et al. Distinct pools of cancer stem-like cells coexist within human glioblastomas and display different tumorigenicity and independent genomic evolution. Oncogene 28, 1807–1811 (2009).
Solit, D. & Sawyers, C. L. How melanomas bypass new therapy. Nature 468, 902–903 (2010).
Goff, D. & Jamieson, C. Cycling toward elimination of leukemic stem cells. Cell Stem Cell 6, 296–297 (2010).
Sharma, S. V. et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80 (2010).
Chin, L., Andersen, J. N. & Futreal, P. A. Cancer genomics: from discovery science to personalized medicine. Nature Med. 17, 297–303 (2011).
Sawyers, C. L. Shifting paradigms: the seeds of oncogene addiction. Nature Med. 15, 1158–1161 (2009).
Graham, S. M. et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro . Blood 99, 319–325 (2002).
Turke, A. B. et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 17, 77–88 (2010).
Ashworth, A., Lord, C. J. & Reis-Filho, J. S. Genetic interactions in cancer progression and treatment. Cell 145, 30–38 (2011).
Zhang, B. et al. Effective targeting of quiescent chronic myelogenous leukemia stem cells by histone deacetylase inhibitors in combination with imatinib mesylate. Cancer Cell 17, 427–442 (2010).
Duy, C. et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR–ABL1 kinase inhibition. Nature 473, 384–388 (2011).
Pienta, K. J., McGregor, N., Axelrod, R. & Axelrod, D. E. Ecological therapy for cancer: defining tumors using an ecosystem paradigm suggests new opportunities for novel cancer treatments. Trans. Oncol. 1, 158–164 10.1593/tlo.08178 (2008).
Calabrese, C. et al. A perivascular niche for brain tumor stem cells. Cancer Cell 11, 69–82 (2007).
Bissell, M. J. & Hines, W. C. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nature Med. 17, 320–329 (2011).
Wargo, A. R., Huijben, S., de Roode, J. C., Shepherd, J. & Read, A. F. Competitive release and facilitation of drug-resistant parasites after therapeutic chemotherapy in a rodent malaria model. Proc. Natl Acad. Sci. USA 104, 19914–19919 (2007).
Gatenby, R. A., Silva, A. S., Gillies, R. J. & Frieden, B. R. Adaptive therapy. Cancer Res. 69, 4894–4903 (2009). Dosing to maintain tumour size prolonged survival far longer than high-dose therapy in a mouse xenograft model.
Acknowledgements
The research of M.G. is supported by Leukaemia & Lymphoma Research UK and The Kay Kendall Leukaemia Fund. The research of C.M. is supported by Research Scholar Grant #117209-RSG-09-163-01-CNE from the American Cancer Society and NIH grants P01 CA91955, U54 CA143803, R01 CA149566 and R01 CA140657. The authors thank C.Cooper and J.Clark for the use of the image in Fig. 4.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reprints and permissions information is available at www.nature.com/reprints.
Supplementary information
Rights and permissions
About this article
Cite this article
Greaves, M., Maley, C. Clonal evolution in cancer. Nature 481, 306–313 (2012). https://doi.org/10.1038/nature10762
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10762
This article is cited by
-
Inferring single-cell copy number profiles through cross-cell segmentation of read counts
BMC Genomics (2024)
-
CONIPHER: a computational framework for scalable phylogenetic reconstruction with error correction
Nature Protocols (2024)
-
Recent developments in targeting breast cancer stem cells (BCSCs): a descriptive review of therapeutic strategies and emerging therapies
Medical Oncology (2024)
-
The Human Digi-real Duality
SN Computer Science (2024)
-
The tumor ecosystem in head and neck squamous cell carcinoma and advances in ecotherapy
Molecular Cancer (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.