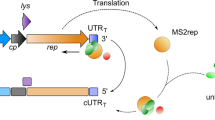
Abstract
Most retroviruses require translational recoding of a viral messenger RNA stop codon to maintain a precise ratio of structural (Gag) and enzymatic (Pol) proteins during virus assembly1,2. Pol is expressed exclusively as a Gag–Pol fusion either by ribosomal frameshifting or by read-through of the gag stop codon3. Both of these mechanisms occur infrequently and only affect 5–10% of translating ribosomes, allowing the virus to maintain the critical Gag to Gag–Pol ratio4,5,6,7,8. Although it is understood that the frequency of the recoding event is regulated by cis RNA motifs, no mechanistic explanation is currently available for how the critical protein ratio is maintained. Here we present the NMR structure of the murine leukaemia virus recoding signal and show that a protonation-dependent switch occurs to induce the active conformation. The equilibrium is such that at physiological pH the active, read-through permissive conformation is populated at approximately 6%: a level that correlates with in vivo protein quantities. The RNA functions by a highly sensitive, chemo-mechanical coupling tuned to ensure an optimal read-through frequency. Similar observations for a frameshifting signal indicate that this novel equilibrium-based mechanism may have a general role in translational recoding.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Felsenstein, K. M. & Goff, S. P. Expression of the Gag-Pol fusion protein of Moloney murine leukemia virus without Gag protein does not induce virion formation or proteolytic processing. J. Virol. 62, 2179–2182 (1988)
Shehu-Xhilaga, M., Crowe, S. M. & Mak, J. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 75, 1834–1841 (2001)
Baranov, P. V., Gesteland, R. F. & Atkins, J. F. Recoding: translational bifurcations in gene expression. Gene 286, 187–201 (2002)
Philipson, L. et al. Translation of MuLV and MSV RNAs in nuclease-treated reticulocyte extracts: enhancement of the Gag-Pol polypeptide with yeast suppressor tRNA. Cell 13, 189–199 (1978)
Yoshinaka, Y., Katoh, I., Copeland, T. D. & Oroszlan, S. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc. Natl Acad. Sci. USA 82, 1618–1622 (1985)
Wills, N. M., Gesteland, R. F. & Atkins, J. F. Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc. Natl Acad. Sci. USA 88, 6991–6995 (1991)
Wills, N. M., Gesteland, R. F. & Atkins, J. F. Pseudoknot-dependent read-through of retroviral gag termination codons: importance of sequences in the spacer and loop 2. EMBO J. 13, 4137–4144 (1994)
Alam, S. L., Wills, N. M., Ingram, J. A., Atkins, J. F. & Gesteland, R. F. Structural studies of the RNA pseudoknot required for readthrough of the gag-termination codon of murine leukemia virus. J. Mol. Biol. 288, 837–852 (1999)
Brierley, I., Pennell, S. & Gilbert, R. J. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nature Rev. Microbiol. 5, 598–610 (2007)
Honigman, A., Wolf, D., Yaish, S., Falk, H. & Panet, A. cis Acting RNA sequences control the gag-pol translation readthrough in murine leukemia virus. Virology 183, 313–319 (1991)
Feng, Y. X., Yuan, H., Rein, A. & Levin, J. G. Bipartite signal for read-through suppression in murine leukemia virus mRNA: an eight-nucleotide purine-rich sequence immediately downstream of the gag termination codon followed by an RNA pseudoknot. J. Virol. 66, 5127–5132 (1992)
Nixon, P. L. & Giedroc, D. P. Energetics of a strongly pH dependent RNA tertiary structure in a frameshifting pseudoknot. J. Mol. Biol. 296, 659–671 (2000)
Grentzmann, G., Ingram, J. A., Kelly, P. J., Gesteland, R. F. & Atkins, J. F. A dual-luciferase reporter system for studying recoding signals. RNA 4, 479–486 (1998)
Schroeder, S., Kim, J. & Turner, D. H. G. A. and U.U mismatches can stabilize RNA internal loops of three nucleotides. Biochemistry 35, 16105–16109 (1996)
Badhwar, J., Karri, S., Cass, C. K., Wunderlich, E. L. & Znosko, B. M. Thermodynamic characterization of RNA duplexes containing naturally occurring 1 × 2 nucleotide internal loops. Biochemistry 46, 14715–14724 (2007)
Michiels, P. J. et al. Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting. J. Mol. Biol. 310, 1109–1123 (2001)
Clanton-Arrowood, K., McGurk, J. & Schroeder, S. J. 3′ terminal nucleotides determine thermodynamic stabilities of mismatches at the ends of RNA helices. Biochemistry 47, 13418–13427 (2008)
Cornish, P. V., Hennig, M. & Giedroc, D. P. A loop 2 cytidine-stem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated −1 ribosomal frameshifting. Proc. Natl Acad. Sci. USA 102, 12694–12699 (2005)
Atkins, J. F. & Gesteland, R. F. Recoding: Expansion of Decoding Rules Enriches Gene Expression (Springer, 2010)
Al-Hashimi, H. M. & Walter, N. G. RNA dynamics: it is about time. Curr. Opin. Struct. Biol. 18, 321–329 (2008)
Namy, O., Moran, S. J., Stuart, D. I., Gilbert, R. J. & Brierley, I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 441, 244–247 (2006)
Giedroc, D. P. & Cornish, P. V. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 139, 193–208 (2009)
Agirrezabala, X. & Frank, J. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q. Rev. Biophys. 42, 159–200 (2009)
D’Souza, V., Dey, A., Habib, D. & Summers, M. F. NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J. Mol. Biol. 337, 427–442 (2004)
Güntert, P., Mumenthaler, C. & Wüthrich, K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 (1997)
Kuhlman, B., Luisi, D. L., Young, P. & Raleigh, D. P. pKa values and the pH dependent stability of the N-terminal domain of L9 as probes of electrostatic interactions in the denatured state. Differentiation between local and nonlocal interactions. Biochemistry 38, 4896–4903 (1999)
Legault, P. & Pardi, A. In-situ probing of adenine protonation in RNA by 13C NMR. J. Am. Chem. Soc. 116, 8390–8391 (1994)
Staple, D. W. & Butcher, S. E. Solution structure and thermodynamic investigation of the HIV-1 frameshift inducing element. J. Mol. Biol. 349, 1011–1023 (2005)
Acknowledgements
We thank J. Atkins for the p2luc plasmid, the New York Structural Biology Center for NMR time and A. Hawkins, S. Leiman and J. Gullinger for their contributions. B.H.-L. and S.P.G. would also like to thank the late D. Wolf for his inspiration and many helpful discussions. S.P.G. acknowledges grant R37 CA30488 from the NCI/NIH and is an Investigator with the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
V.M.D’S., B.H.-L., M.A.D. and S.P.G. conceived of and designed the experiments. V.M.D’S., M.A.D., C.S., N.S. and B.H.-L. did the structural analysis, B.H.-L. did the recoding assays, and M.A.D. and J.M.N. did the in vivo assay for the double mutant. M.A.D. and V.M.D’S. wrote the manuscript with assistance from B.H.-L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Text, Supplementary References, Supplementary Table 1 and Supplementary Figures 1-14 with legends. (PDF 7353 kb)
Rights and permissions
About this article
Cite this article
Houck-Loomis, B., Durney, M., Salguero, C. et al. An equilibrium-dependent retroviral mRNA switch regulates translational recoding. Nature 480, 561–564 (2011). https://doi.org/10.1038/nature10657
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10657
This article is cited by
-
Length-dependent motions of SARS-CoV-2 frameshifting RNA pseudoknot and alternative conformations suggest avenues for frameshifting suppression
Nature Communications (2022)
-
Engineered pegRNAs improve prime editing efficiency
Nature Biotechnology (2022)
-
Visualizing a protonated RNA state that modulates microRNA-21 maturation
Nature Chemical Biology (2021)
-
QRNAS: software tool for refinement of nucleic acid structures
BMC Structural Biology (2019)
-
Viral RNA structure-based strategies to manipulate translation
Nature Reviews Microbiology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.