Abstract
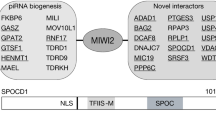
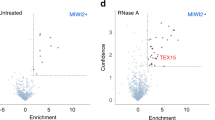
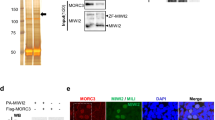
Piwi proteins and Piwi-interacting RNAs (piRNAs) have conserved functions in transposon silencing1. The murine Piwi proteins Mili and Miwi2 (also called Piwil2 and Piwil4, respectively) direct epigenetic LINE1 and intracisternal A particle transposon silencing during genome reprogramming in the embryonic male germ line2,3,4. Piwi proteins are proposed to be piRNA-guided endonucleases that initiate secondary piRNA biogenesis5,6,7; however, the actual contribution of their endonuclease activities to piRNA biogenesis and transposon silencing remain unknown. To investigate the role of Piwi-catalysed endonucleolytic activity, we engineered point mutations in mice that substitute the second aspartic acid to an alanine in the DDH catalytic triad of Mili and Miwi2, generating the MiliDAH and Miwi2DAH alleles, respectively. Analysis of Mili-bound piRNAs from homozygous MiliDAH fetal gonadocytes revealed a failure of transposon piRNA amplification, resulting in the marked reduction of piRNA bound within Miwi2 ribonuclear particles. We find that Mili-mediated piRNA amplification is selectively required for LINE1, but not intracisternal A particle, silencing. The defective piRNA pathway in MiliDAH mice results in spermatogenic failure and sterility. Surprisingly, homozygous Miwi2DAH mice are fertile, transposon silencing is established normally and no defects in secondary piRNA biogenesis are observed. In addition, the hallmarks of piRNA amplification are observed in Miwi2-deficient gonadocytes. We conclude that cycles of intra-Mili secondary piRNA biogenesis fuel piRNA amplification that is absolutely required for LINE1 silencing.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
ArrayExpress
Data deposits
All raw sequencing data are deposited in ArrayExpress (accession number E-MTAB-730) and European Nucleotide Archive (ERP000778). The MiliDAH, Miwi2DAH and Miwi2 null (Miwi2−) alleles have been deposited at EMMA (http://www.emmanet.org/) and will be freely available on a non-collaborative basis.
Change history
30 October 2011
The ArrayExpress data accession number was corrected.
References
Ghildiyal, M. & Zamore, P. D. Small silencing RNAs: an expanding universe. Nature Rev. Genet. 10, 94–108 (2009)
Aravin, A. A., Sachidanandam, R., Girard, A., Fejes-Toth, K. & Hannon, G. J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744–747 (2007)
Carmell, M. A. et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12, 503–514 (2007)
Kuramochi-Miyagawa, S. et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22, 908–917 (2008)
Brennecke, J. et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103 (2007)
Gunawardane, L. S. et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315, 1587–1590 (2007)
Aravin, A. A. et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799 (2008)
Song, J. J., Smith, S. K., Hannon, G. J. & Joshua-Tor, L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305, 1434–1437 (2004)
Patel, D. J. et al. Structural biology of RNA silencing and its functional implications. Cold Spring Harb. Symp. Quant. Biol. 71, 81–93 (2006)
Saito, K. et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20, 2214–2222 (2006)
Bestor, T. H. & Bourc’his, D. Transposon silencing and imprint establishment in mammalian germ cells. Cold Spring Harb. Symp. Quant. Biol. 69, 381–388 (2004)
Cheloufi, S., Dos Santos, C. O., Chong, M. M. & Hannon, G. J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465, 584–589 (2010)
Cifuentes, D. et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 328, 1694–1698 (2010)
O’Carroll, D. et al. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 21, 1999–2004 (2007)
Liu, J. et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441 (2004)
Maniataki, E. & Mourelatos, Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 19, 2979–2990 (2005)
Aravin, A. A. et al. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 5, e1000764 (2009)
Kojima, K. et al. Associations between PIWI proteins and TDRD1/MTR-1 are critical for integrated subcellular localization in murine male germ cells. Genes Cells 14, 1155–1165 (2009)
Reuter, M. et al. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nature Struct. Mol. Biol. 16, 639–646 (2009)
Vagin, V. V. et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 23, 1749–1762 (2009)
Wang, J., Saxe, J. P., Tanaka, T., Chuma, S. & Lin, H. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr. Biol. 19, 640–644 (2009)
Zheng, K. et al. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc. Natl Acad. Sci. USA 107, 11841–11846 (2010)
Chuma, S. et al. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc. Natl Acad. Sci. USA 103, 15894–15899 (2006)
Kuramochi-Miyagawa, S. et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 24, 887–892 (2010)
Kuramochi-Miyagawa, S. et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131, 839–849 (2004)
Elbashir, S. M., Lendeckel, W. & Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200 (2001)
Martinez, J. & Tuschl, T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 18, 975–980 (2004)
Mandal, P. K. & Kazazian, H. H., Jr SnapShot: Vertebrate transposons. Cell 135, 192–192 (2008)
Wen, J. & Brogna, S. Nonsense-mediated mRNA decay. Biochem. Soc. Trans. 36, 514–516 (2008)
Robanus-Maandag, E. et al. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev. 12, 1599–1609 (1998)
Poueymirou, W. T. et al. F0 generation mice fully derived from gene-targeted embryonic stem cells allowing immediate phenotypic analyses. Nature Biotechnol. 25, 91–99 (2007)
Farley, F. W., Soriano, P., Steffen, L. S. & Dymecki, S. M. Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28, 106–110 (2000)
Schwenk, F., Baron, U. & Rajewsky, K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23, 5080–5081 (1995)
Yoshimizu, T. et al. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev. Growth Differ. 41, 675–684 (1999)
Hafner, M. et al. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44, 3–12 (2008)
Gentleman, R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004)
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Flicek, P. et al. Ensembl 2011. Nucleic Acids Res. 39, D800–D806 (2011)
Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J. & Wheeler, D. L. GenBank. Nucleic Acids Res. 36, D25–D30 (2008)
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009)
Acknowledgements
We are grateful to R. Pillai, B. Cullen, S. Martin, S. Chuma and J. Lykke-Andersen for antibodies used in this study. This study was technically supported by of EMBL’s genomic core facility. We are very grateful to V. Benes, R. Pillai and S. van Dongen for advice. We are grateful to M. Reuter for assistance with the preparation of immunoprecipitations for mass spectroscopy. This study was technically supported by EMBL Monterotondo’s FACS and Microscopy core facilities. We are grateful to A. Wutz for A9 ES cells. We also acknowledge the services of J. Rientjes from Monash University’s Gene Recombineering Facility. We are also very grateful to C. Kutter and D. Odom for advice on small RNA library generation.
Author information
Authors and Affiliations
Contributions
S.D.F. contributed to the design, execution and analysis of the majority of experiments on MiliDAH and Miwi2DAH mice. N.B. performed the bioinformatic analysis presented in the manuscript with initial assistance from C.A.-G. M.D.G. analysed the spermatogenic defects as well as undertook the co-localization studies in the respective mouse strains. A.S. performed the bisulphite sequencing experiments. C.F. and C.A. performed the electron microscopy experiments. P.N.M. established the 8-cell embryo ES cell injection procedure. A.J.E. supervised the bioinformatic analysis. D.O’C. conceived and supervised this study and wrote the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-16 with legends and Supplementary Tables 1-2. (PDF 8062 kb)
Rights and permissions
About this article
Cite this article
De Fazio, S., Bartonicek, N., Di Giacomo, M. et al. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480, 259–263 (2011). https://doi.org/10.1038/nature10547
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10547
This article is cited by
-
Themes and variations on piRNA-guided transposon control
Mobile DNA (2023)
-
The epigenetic regulatory mechanism of PIWI/piRNAs in human cancers
Molecular Cancer (2023)
-
Emerging roles and functional mechanisms of PIWI-interacting RNAs
Nature Reviews Molecular Cell Biology (2023)
-
Temperature sensitivity of DNA double-strand break repair underpins heat-induced meiotic failure in mouse spermatogenesis
Communications Biology (2022)
-
GTSF1 accelerates target RNA cleavage by PIWI-clade Argonaute proteins
Nature (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.