Abstract
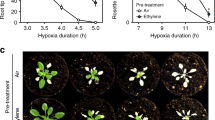
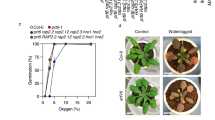
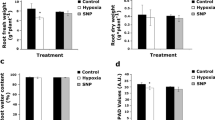
Plants and animals are obligate aerobes, requiring oxygen for mitochondrial respiration and energy production. In plants, an unanticipated decline in oxygen availability (hypoxia), as caused by roots becoming waterlogged or foliage submergence, triggers changes in gene transcription and messenger RNA translation that promote anaerobic metabolism and thus sustain substrate-level ATP production1. In contrast to animals2, oxygen sensing has not been ascribed to a mechanism of gene regulation in response to oxygen deprivation in plants. Here we show that the N-end rule pathway of targeted proteolysis acts as a homeostatic sensor of severe low oxygen levels in Arabidopsis, through its regulation of key hypoxia-response transcription factors. We found that plants lacking components of the N-end rule pathway constitutively express core hypoxia-response genes and are more tolerant of hypoxic stress. We identify the hypoxia-associated ethylene response factor group VII transcription factors of Arabidopsis as substrates of this pathway. Regulation of these proteins by the N-end rule pathway occurs through a characteristic conserved motif at the amino terminus initiating with Met-Cys. Enhanced stability of one of these proteins, HRE2, under low oxygen conditions improves hypoxia survival and reveals a molecular mechanism for oxygen sensing in plants via the evolutionarily conserved N-end rule pathway. SUB1A-1, a major determinant of submergence tolerance in rice3, was shown not to be a substrate for the N-end rule pathway despite containing the N-terminal motif, indicating that it is uncoupled from N-end rule pathway regulation, and that enhanced stability may relate to the superior tolerance of Sub1 rice varieties to multiple abiotic stresses4.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Data deposits
The microarray data reported in this paper are deposited in Gene Expression Omnibus under accession number GSE29941 and are also tabulated in Supplementary Information.
References
Bailey-Serres, J. & Voesenek, L. A. C. J. Flooding stress: Acclimations and genetic diversity. Annu. Rev. Plant Biol. 59, 313–339 (2008)
Kaelin, W. G. & Ratcliffe, P. J. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402 (2008)
Xu, K. et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442, 705–708 (2006)
Fukao, T., Yeung, E. & Bailey-Serres, J. The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23, 412–427 (2011)
Bachmair, A., Finley, D. & Varshavsky, A. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234, 179–186 (1986)
Varshavsky, A. Regulated protein degradation. Trends Biochem. Sci. 30, 283–286 (2005)
Kwon, Y. T. et al. An essential role of N-terminal arginylation in cardiovascular development. Science 297, 96–99 (2002)
Graciet, E., Mesiti, F. & Wellmer, F. Structure and evolutionary conservation of the plant N-end rule pathway. Plant J. 61, 741–751 (2010)
Graciet, E. & Wellmer, F. The plant N-end rule pathway: structure and functions. Trends Plant Sci. 15, 447–453 (2010)
Garzón, M. et al. PRT6/At5g02310 encodes an Arabidopsis ubiquitin ligase of the N-end rule pathway with arginine specificity and is not the CER3 locus. FEBS Lett. 581, 3189–3196 (2007)
Tasaki, T. & Kwon, Y. T. The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem. Sci. 32, 520–528 (2007)
Holman, T. J. et al. The N-end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis . Proc. Natl Acad. Sci. USA 106, 4549–4554 (2009)
Yoshida, S., Ito, M., Callis, J., Nishida, I. & Watanabe, A. A delayed leaf senescence mutant is defective in arginyl-tRNA: protein arginyltransferase, a component of the N-end rule pathway in Arabidopsis . Plant J. 32, 129–137 (2002)
Graciet, E. et al. The N-end rule pathway controls multiple functions during Arabidopsis shoot and leaf development. Proc. Natl Acad. Sci. USA 106, 13618–13623 (2009)
Mustroph, A. et al. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis . Proc. Natl Acad. Sci. USA 106, 18843–18848 (2009)
Chung, H. J. & Ferl, R. J. Arabidopsis alcohol dehydrogenase expression in both shoots and roots is conditioned by root growth environment. Plant Physiol. 121, 429–436 (1999)
Nakano, T., Suzuki, K., Fujimura, T. & Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432 (2006)
Licausi, F. et al. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana . Plant J. 62, 302–315 (2010)
Hinz, M. et al. Arabidopsis RAP2.2: An ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 153, 757–772 (2010)
Papdi, C. et al. Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol. 147, 528–542 (2008)
Hattori, Y. et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460, 1026–1030 (2009)
Hu, R. G. et al. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 437, 981–986 (2005)
Lee, M. J. et al. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc. Natl Acad. Sci. USA 102, 15030–15035 (2005)
Takahashi, H. et al. A simple and high-sensitivity method for analysis of ubiquitination and polyubiquitination based on wheat cell-free protein synthesis. BMC Plant Biol. 9, 39 (2009)
Suzuki, T. & Varshavsky, A. Degradation signals in the lysine-asparagine sequence space. EMBO J. 18, 6017–6026 (1999)
Leonard, S. E. & Carroll, K. S. Chemical ‘omics’ approaches for understanding protein cysteine oxidation in biology. Curr. Opin. Chem. Biol. 15, 88–102 (2011)
Felle, H. H. pH regulation in anoxic plants. Ann. Bot. 96, 519–532 (2005)
Hu, R. G., Wang, H. Q., Xia, Z. X. & Varshavsky, A. The N-end rule pathway is a sensor of heme. Proc. Natl Acad. Sci. USA 105, 76–81 (2008)
Côme, D. & Tissaoui, T. Induction d’une dormance embryonnaire secondaire chez le pommier (Pirus malus L.) par des atmosphères très appauvries en oxygène. Compt. Rendus Acad. Sci. 266, 477–479 (1968)
Branco-Price, C., Kaiser, K. A., Jang, C. J. H., Larive, C. K. & Bailey-Serres, J. Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana . Plant J. 56, 743–755 (2008)
Mustroph, A. et al. Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 152, 1484–1500 (2010)
Lucas, M. et al. SHORT-ROOT regulates primary, lateral, and adventitious root development in Arabidopsis . Plant Physiol. 155, 384–398 (2011)
Dubin, M. J., Bowler, C. & Benvenuto, G. A modified Gateway cloning strategy for overexpressing tagged proteins in plants. Plant Methods 4, 3 (2008)
Swarup, R. et al. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nature Cell Biol. 7, 1057–1065 (2005)
Martinez-Garcia, J. F., Monte, E. & Quail, P. H. A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20, 251–257 (1999)
Williams, A. J., Werner-Fraczek, J., Chang, I. F. & Bailey-Serres, J. Regulated phosphorylation of 40S ribosomal protein S6 in root tips of maize. Plant Physiol. 132, 2086–2097 (2003)
Thompson, J. D., Higgins, D. G. & Gibson, T. J. Clustal-W - Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994)
Horan, K. et al. Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol. 147, 41–57 (2008)
Acknowledgements
M.J.H., D.J.G., S.G. and C.S.C. were supported by BBSRC grant BB/G010595/1; G.W.B. by a Marie Curie International Incoming Fellowship; N.M.I. by a MARA PhD fellowship from the Malaysian government; S.C.L., T.F. and J.B.-S. by grants NSF IOS-0750811 and NIFA 2008-35100-04528. We thank S. Liddell. Rothamsted Research receives grant-aided support from the BBSRC.
Author information
Authors and Affiliations
Contributions
D.J.G., M.J.H., J.B.-S., F.C. and F.L.T. conceived and designed experiments. D.J.G., S.C.L., N.M.I., S.G., C.S.C., G.W.B., T.F. and F.C. performed the experiments. D.J.G., S.C.L., N.M.I., S.G., C.S.C., G.W.B., T.F., F.C. M.J.H., J.B-S. and F.L.T. analysed the data. M.J.H., D.J.G. and J.B.-S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
The file contains Supplementary Figures 1-5 with legends. (PDF 718 kb)
Supplementary Table 1
This file contains microarray data comparing gene expression in Col-0 (WT), prt6-1 and ate1-2 ate2-1 mutants. (XLS 310 kb)
Supplementary Table 2
This file contains a list of oligonucleotide primers used in this study. (XLS 14 kb)
Rights and permissions
About this article
Cite this article
Gibbs, D., Lee, S., Md Isa, N. et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479, 415–418 (2011). https://doi.org/10.1038/nature10534
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10534
This article is cited by
-
ERF111/ABR1: An AP2 Domain Transcription Factor Serving as a Hub for Multiple Signaling Pathways
Journal of Plant Growth Regulation (2024)
-
A microscopic scenario on recovery mechanisms under waterlogging and submergence stress in rice
Planta (2024)
-
Navigating Through Harsh Conditions: Coordinated Networks of Plant Adaptation to Abiotic Stress
Journal of Plant Growth Regulation (2024)
-
ERFVII action and modulation through oxygen-sensing in Arabidopsis thaliana
Nature Communications (2023)
-
Iron-sulfur clusters are involved in post-translational arginylation
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.