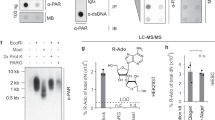
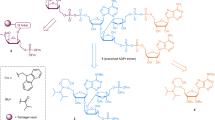
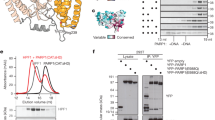
Abstract
Post-translational modification of proteins by poly(ADP-ribosyl)ation regulates many cellular pathways that are critical for genome stability, including DNA repair, chromatin structure, mitosis and apoptosis1. Poly(ADP-ribose) (PAR) is composed of repeating ADP-ribose units linked via a unique glycosidic ribose–ribose bond, and is synthesized from NAD by PAR polymerases1,2. PAR glycohydrolase (PARG) is the only protein capable of specific hydrolysis of the ribose–ribose bonds present in PAR chains; its deficiency leads to cell death3,4. Here we show that filamentous fungi and a number of bacteria possess a divergent form of PARG that has all the main characteristics of the human PARG enzyme. We present the first PARG crystal structure (derived from the bacterium Thermomonospora curvata), which reveals that the PARG catalytic domain is a distant member of the ubiquitous ADP-ribose-binding macrodomain family5,6. High-resolution structures of T. curvata PARG in complexes with ADP-ribose and the PARG inhibitor ADP-HPD, complemented by biochemical studies, allow us to propose a model for PAR binding and catalysis by PARG. The insights into the PARG structure and catalytic mechanism should greatly improve our understanding of how PARG activity controls reversible protein poly(ADP-ribosyl)ation and potentially of how the defects in this regulation are linked to human disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hakmé, A., Wong, H. K., Dantzer, F. & Schreiber, V. The expanding field of poly(ADP-ribosyl)ation reactions. EMBO Rep. 9, 1094–1100 (2008)
D’Amours, D., Desnoyers, S., D’Silva, I. & Poirier, G. G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342, 249–268 (1999)
Koh, D. W. et al. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl Acad. Sci. USA 101, 17699–17704 (2004)
Hanai, S. et al. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster . Proc. Natl Acad. Sci. USA 101, 82–86 (2004)
Karras, G. I. et al. The macro domain is an ADP-ribose binding module. EMBO J. 24, 1911–1920 (2005)
Till, S. & Ladurner, A. G. Sensing NAD metabolites through macro domains. Front. Biosci. 14, 3246–3258 (2009)
Patel, C. N., Koh, D. W., Jacobson, M. K. & Oliveira, M. A. Identification of three critical acidic residues of poly(ADP-ribose) glycohydrolase involved in catalysis: determining the PARG catalytic domain. Biochem. J. 388, 493–500 (2005)
Panda, S., Poirier, G. G. & Kay, S. A. tej defines a role for poly(ADP-ribosyl)ation in establishing period length of the Arabidopsis circadian oscillator. Dev. Cell 3, 51–61 (2002)
Koh, D. W. et al. Identification of an inhibitor binding site of poly(ADP-ribose) glycohydrolase. Biochemistry 42, 4855–4863 (2003)
Ahel, I. et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451, 81–85 (2008)
Kothe, G. O., Kitamura, M., Masutani, M., Selker, E. U. & Inoue, H. PARP is involved in replicative aging in Neurospora crassa . Fungal Genet. Biol. 47, 297–309 (2010)
Semighini, C. P., Savoldi, M., Goldman, G. H. & Harris, S. D. Functional characterization of the putative Aspergillus nidulans poly(ADP-ribose) polymerase homolog PrpA. Genetics 173, 87–98 (2006)
Hassa, P. O. & Hottiger, M. O. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 13, 3046–3082 (2008)
Tao, Z., Gao, P. & Liu, H. W. Studies of the expression of human poly(ADP-ribose) polymerase-1 in Saccharomyces cerevisiae and identification of PARP-1 substrates by yeast proteome microarray screening. Biochemistry 48, 11745–11754 (2009)
Lin, W., Ame, J. C., Aboul-Ela, N., Jacobson, E. L. & Jacobson, M. K. Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 272, 11895–11901 (1997)
Kustatscher, G., Hothorn, M., Pugieux, C., Scheffzek, K. & Ladurner, A. G. Splicing regulates NAD metabolite binding to histone macroH2A. Nature Struct. Mol. Biol. 12, 624–625 (2005)
Ahel, D. et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325, 1240–1243 (2009)
Gottschalk, A. J. et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl Acad. Sci. USA 106, 13770–13774 (2009)
Timinszky, G. et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nature Struct. Mol. Biol. 16, 923–929 (2009)
Mueller-Dieckmann, C. et al. The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc. Natl Acad. Sci. USA 103, 15026–15031 (2006)
Oka, S., Kato, J. & Moss, J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 281, 705–713 (2006)
Alberti, S., Gitler, A. D. & Lindquist, S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae . Yeast 24, 913–919 (2007)
Kabsch, W. Evaluation of single-crystal X-ray diffraction data from a position-sensitive detector. J. Appl. Cryst. 21, 916–924 (1988)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Perrakis, A., Morris, R. & Lamzin, V. S. Automated protein model building combined with iterative structure refinement. Nature Struct. Biol. 6, 458–463 (1999)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005)
Cornell, W. D. et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117, 5179–5197 (1995)
Jakalian, A., Jack, D. B. & Bayly, C. I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 23, 1623–1641 (2002)
Coulier, L. et al. Simultaneous quantitative analysis of metabolites using ion-pair liquid chromatography-electrospray ionization mass spectrometry. Anal. Chem. 78, 6573–6582 (2006)
Acknowledgements
We thank G. Clark for genomic DNA from E. dispar, G. Smith for the PARP inhibitor, M. Rossi for purified proteins and R. Thorough for editing English. We thank D. Ahel, A. Jordan, D. Ogilvie, S. Terzic, D. Neuhaus and S. Eustermann for helpful discussions. We are grateful to B. Lüscher for the gift of the PARP10 expression plasmid, and K. Labib and the members of his laboratory for advice with yeast work. This work was funded by Cancer Research UK. D.S. holds an AXA Research Fund post-doctoral fellowship. D.L. is a Royal Society University Research Fellow. Access to Diamond beamlines is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
D.S. performed biochemical and in vivo experiments, prepared proteins for crystallization, analysed data and wrote the manuscript. M.S.D. performed structural/biophysical studies and analysed data. E.B. performed biochemical and in vivo experiments; R.W. performed supporting studies. M.A. and N.D. performed LC/MS analyses; P.L. performed molecular modelling studies. I.A. and D.L. wrote the manuscript, designed experiments and analysed data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains a Supplementary Discussion, Supplementary Table 1, Supplementary Figures 1-9 with legends and Supplementary References. (PDF 775 kb)
Rights and permissions
About this article
Cite this article
Slade, D., Dunstan, M., Barkauskaite, E. et al. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature 477, 616–620 (2011). https://doi.org/10.1038/nature10404
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10404
This article is cited by
-
Serine ADP-ribosylation in Drosophila provides insights into the evolution of reversible ADP-ribosylation signalling
Nature Communications (2023)
-
Poly(ADP-ribosylation) of P-TEFb by PARP1 disrupts phase separation to inhibit global transcription after DNA damage
Nature Cell Biology (2022)
-
Functions of ADP-ribose transferases in the maintenance of telomere integrity
Cellular and Molecular Life Sciences (2022)
-
Serine-linked PARP1 auto-modification controls PARP inhibitor response
Nature Communications (2021)
-
Mechanistic insights into the three steps of poly(ADP-ribosylation) reversal
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.