Abstract
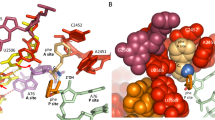
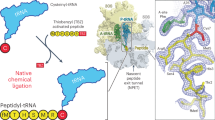
The active site of the ribosome, the peptidyl transferase centre, catalyses two reactions, namely, peptide bond formation between peptidyl-tRNA and aminoacyl-tRNA as well as the release-factor-dependent hydrolysis of peptidyl-tRNA. Unlike peptide bond formation, peptide release is strongly impaired by mutations of nucleotides within the active site, in particular by base exchanges at position A2602 (refs 1, 2). The 2′-OH group of A76 of the peptidyl - tRNA substrate seems to have a key role in peptide release3. According to computational analysis4 , the 2′-OH may take part in a concerted ‘proton shuttle’ by which the leaving group is protonated, in analogy to similar current models of peptide bond formation4,5,6. Here we report kinetic solvent isotope effects and proton inventories (reaction rates measured in buffers with increasing content of deuterated water, D2O) of the two reactions catalysed by the active site of the Escherichia coli ribosome. The transition state of the release factor 2 (RF2)-dependent hydrolysis reaction is characterized by the rate-limiting formation of a single strong hydrogen bond. This finding argues against a concerted proton shuttle in the transition state of the hydrolysis reaction. In comparison, the proton inventory for peptide bond formation indicates the rate-limiting formation of three hydrogen bonds with about equal contributions, consistent with a concerted eight-membered proton shuttle in the transition state5. Thus, the ribosome supports different rate-limiting transition states for the two reactions that take place in the peptidyl transferase centre.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Polacek, N. et al. The critical role of the universally conserved A2602 of 23S ribosomal RNA in the release of the nascent peptide during translation termination. Mol. Cell 11, 103–112 (2003)
Youngman, E. M., Brunelle, J. L., Kochaniak, A. B. & Green, R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117, 589–599 (2004)
Brunelle, J. L., Shaw, J. J., Youngman, E. M. & Green, R. Peptide release on the ribosome depends critically on the 2' OH of the peptidyl-tRNA substrate. RNA 14, 1526–1531 (2008)
Trobro, S. & Aqvist, J. Mechanism of the translation termination reaction on the ribosome. Biochemistry 48, 11296–11303 (2009)
Wallin, G. & Aqvist, J. The transition state for peptide bond formation reveals the ribosome as a water trap. Proc. Natl Acad. Sci. USA 107, 1888–1893 (2010)
Jin, H., Kelley, A. C., Loakes, D. & Ramakrishnan, V. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc. Natl Acad. Sci. USA 107, 8593–8598 (2010)
Laurberg, M. et al. Structural basis for translation termination on the 70S ribosome. Nature 454, 852–857 (2008)
Weixlbaumer, A. et al. Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322, 953–956 (2008)
Korostelev, A. et al. Crystal structure of a translation termination complex formed with release factor RF2. Proc. Natl Acad. Sci. USA 105, 19684–19689 (2008)
Petry, S. et al. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 123, 1255–1266 (2005)
Schmeing, T. M., Huang, K. S., Kitchen, D. E., Strobel, S. A. & Steitz, T. A. Structural insights into the roles of water and the 2' hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol. Cell 20, 437–448 (2005)
Shaw, J. J. & Green, R. Two distinct components of release factor function uncovered by nucleophile partitioning analysis. Mol. Cell 28, 458–467 (2007)
Trobro, S. & Aqvist, J. Analysis of predictions for the catalytic mechanism of ribosomal peptidyl transfer. Biochemistry 45, 7049–7056 (2006)
Quinn, D. M. in Isotope Effects in Chemistry and Biology (eds Kohen, A. & Limbach, H. H. ) 995–1018 (CRC Taylor and Francis, 2006)
Schowen, K. B. J. in Transition States of Biochemical Processes (eds Gandour, R. D. & Schowen, R. L. ) 225–283 (Plenum, 1972)
Amort, M. et al. An intact ribose moiety at A2602 of 23S rRNA is key to trigger peptidyl-tRNA hydrolysis during translation termination. Nucleic Acids Res. 35, 5130–5140 (2007)
Mora, L. et al. The essential role of the invariant GGQ motif in the function and stability in vivo of bacterial release factors RF1 and RF2. Mol. Microbiol. 47, 267–275 (2003)
Katunin, V. I., Muth, G. W., Strobel, S. A., Wintermeyer, W. & Rodnina, M. V. Important contribution to catalysis of peptide bond formation by a single ionizing group within the ribosome. Mol. Cell 10, 339–346 (2002)
Kingery, D. A. et al. An uncharged amine in the transition state of the ribosomal peptidyl transfer reaction. Chem. Biol. 15, 493–500 (2008)
Seila, A. C., Okuda, K., Nunez, S., Seila, A. F. & Strobel, S. A. Kinetic isotope effect analysis of the ribosomal peptidyl transferase reaction. Biochemistry 44, 4018–4027 (2005)
Beringer, M., Adio, S., Wintermeyer, W. & Rodnina, M. The G2447A mutation does not affect ionization of a ribosomal group taking part in peptide bond formation. RNA 9, 919–922 (2003)
Beringer, M. et al. Essential mechanisms in the catalysis of peptide bond formation on the ribosome. J. Biol. Chem. 280, 36065–36072 (2005)
Bieling, P., Beringer, M., Adio, S. & Rodnina, M. V. Peptide bond formation does not involve acid-base catalysis by ribosomal residues. Nature Struct. Mol. Biol. 13, 423–428 (2006)
Huang, K. S., Carrasco, N., Pfund, E. & Strobel, S. A. Transition state chirality and role of the vicinal hydroxyl in the ribosomal peptidyl transferase reaction. Biochemistry 47, 8822–8827 (2008)
Rangelov, M. A., Vayssilov, G. N., Yomtova, V. M. & Petkov, D. D. The syn-oriented 2-OH provides a favorable proton transfer geometry in 1,2-diol monoester aminolysis: implications for the ribosome mechanism. J. Am. Chem. Soc. 128, 4964–4965 (2006)
Sievers, A., Beringer, M., Rodnina, M. V. & Wolfenden, R. The ribosome as an entropy trap. Proc. Natl Acad. Sci. USA 101, 7897–7901 (2004)
Dincbas-Renqvist, V. et al. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 19, 6900–6907 (2000)
Freistroffer, D. V., Kwiatkowski, M., Buckingham, R. H. & Ehrenberg, M. The accuracy of codon recognition by polypeptide release factors. Proc. Natl Acad. Sci. USA 97, 2046–2051 (2000)
Glasoe, P. K. & Long, F. A. Use of glass electrodes to measure acidities in deuterium oxide. J. Phys. Chem. 64, 188–190 (1960)
Acknowledgements
We thank H.-H. Limbach for discussions and advice. This work was supported by the Deutsche Forschungsgemeinschaft (M.V.R. and W.W.).
Author information
Authors and Affiliations
Contributions
S.K., W.W. and M.V.R. conceived the study and designed experiments. S.K. performed experiments. All three authors discussed results and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Table 1, additional references and Supplementary Figure 1-2 with legends. (PDF 189 kb)
Rights and permissions
About this article
Cite this article
Kuhlenkoetter, S., Wintermeyer, W. & Rodnina, M. Different substrate-dependent transition states in the active site of the ribosome. Nature 476, 351–354 (2011). https://doi.org/10.1038/nature10247
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10247
This article is cited by
-
Insights into the ribosome function from the structures of non-arrested ribosome–nascent chain complexes
Nature Chemistry (2023)
-
On the Origin of Sugar Handedness: Facts, Hypotheses and Missing Links-A Review
Origins of Life and Evolution of Biospheres (2022)
-
Mechanism of ribosome rescue by alternative ribosome-rescue factor B
Nature Communications (2020)
-
Conformation of methylated GGQ in the Peptidyl Transferase Center during Translation Termination
Scientific Reports (2018)
-
An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome
Nature Structural & Molecular Biology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.