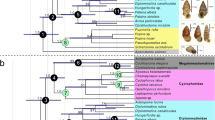
Abstract
Annelida, the ringed worms, is a highly diverse animal phylum that includes more than 15,000 described species and constitutes the dominant benthic macrofauna from the intertidal zone down to the deep sea. A robust annelid phylogeny would shape our understanding of animal body-plan evolution and shed light on the bilaterian ground pattern. Traditionally, Annelida has been split into two major groups: Clitellata (earthworms and leeches) and polychaetes (bristle worms), but recent evidence suggests that other taxa that were once considered to be separate phyla (Sipuncula, Echiura and Siboglinidae (also known as Pogonophora)) should be included in Annelida1,2,3,4. However, the deep-level evolutionary relationships of Annelida are still poorly understood, and a robust reconstruction of annelid evolutionary history is needed. Here we show that phylogenomic analyses of 34 annelid taxa, using 47,953 amino acid positions, recovered a well-supported phylogeny with strong support for major splits. Our results recover chaetopterids, myzostomids and sipunculids in the basal part of the tree, although the position of Myzostomida remains uncertain owing to its long branch. The remaining taxa are split into two clades: Errantia (which includes the model annelid Platynereis), and Sedentaria (which includes Clitellata). Ancestral character trait reconstructions indicate that these clades show adaptation to either an errant or a sedentary lifestyle, with alteration of accompanying morphological traits such as peristaltic movement, parapodia and sensory perception. Finally, life history characters in Annelida seem to be phylogenetically informative.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
Data deposits
Sequence data have been deposited in the NCBI Expressed Sequence Tag database (dbEST) under accession numbers FN424437–FN428571, FR754554–FR771822, HQ729923–HQ729975. The largest aligned data set has been deposited at http://www.treebase.org.
References
Dordel, J., Fisse, F., Purschke, G. & Struck, T. H. Phylogenetic position of Sipuncula derived from multi-gene and phylogenomic data and its implication for the evolution of segmentation. J. Zool. Syst. Evol. Res. 48, 197–207 (2010)
Struck, T. H., Nesnidal, M. P., Purschke, G. & Halanych, K. M. Detecting possibly saturated positions in 18S and 28S sequences and their influence on phylogenetic reconstruction of Annelida (Lophotrochozoa). Mol. Phylogenet. Evol. 48, 628–645 (2008)
Struck, T. H. et al. Annelida phylogeny and the status of Sipuncula and Echiura. BMC Evol. Biol. 7, 57 (2007)
McHugh, D. Molecular evidence that echiurans and pogonophorans are derived annelids. Proc. Natl Acad. Sci. USA 94, 8006–8009 (1997)
Raible, F. et al. Vertebrate-type intron-rich genes in the marine annelid Platynereis dumerilii. Science 310, 1325–1326 (2005)
Tessmar-Raible, K. & Arendt, D. Emerging systems: between vertebrates and arthropods, the Lophotrochozoa. Curr. Opin. Genet. Dev. 13, 331–340 (2003)
Rivera, A. & Weisblat, D. And Lophotrochozoa makes three: Notch/Hes signaling in annelid segmentation. Dev. Genes Evol. 219, 37–43 (2009)
Shain, D. H. Annelids in Modern Biology (Wiley, 2009)
Erséus, C. Phylogeny of oligochaetous Clitellata. Hydrobiologia 535–536, 357–372 (2005)
McHugh, D. Molecular systematics of polychaetes (Annelida). Hydrobiologia 535–536, 309–318 (2005)
Fauvel, P. Polychètes errantes. Faune de France 5, 1–488 (1923)
Fauvel, P. Polychètes sédentaires. Faune de France 16, 1–494 (1927)
de Quatrefages, A. M. Histoire Naturelle des Annelides, Marine et d'Eau Douce. Annelides et Gephyriens Vol. 1 (Librairie Encyclopédique de Roret, 1866)
Day, J. H. A Monograph on the Polychaeta of Southern Africa. Part 1. Errantia (British Museum (Natural History), 1967)
Rouse, G. W. & Fauchald, K. Cladistics and polychaetes. Zool. Scr. 26, 139–204 (1997)
Eibye-Jacobsen, D. A reevaluation of Wiwaxia and the polychaetes of the Burgess Shale. Lethaia 37, 317–335 (2004)
Rouse, G. W. & Pleijel, F. Polychaetes (Oxford Univ. Press, 2001)
Bleidorn, C. et al. On the phylogenetic position of Myzostomida: can 77 genes get it wrong? BMC Evol. Biol. 9, 150 (2009)
Bleidorn, C. et al. Mitochondrial genome and nuclear sequence data support Myzostomida as part of the annelid radiation. Mol. Biol. Evol. 24, 1690–1701 (2007)
Eeckhaut, I., Fievez, L. & Müller, M. C. M. Larval development of Myzostoma cirriferum (Myzostomida). J. Morphol. 258, 269–283 (2003)
Westheide, W. The direction of evolution within the Polychaeta. J. Nat. Hist. 31, 1–15 (1997)
Suschenko, D. & Purschke, G. Ultrastructure of pigmented adult eyes in errant polychaetes (Annelida): implications for annelid evolution. Zoomorphology 128, 75–96 (2009)
Fauchald, K. & Jumars, P. A. The diet of worms: a study of polychaete feedings guilds. Oceanogr. Mar. Biol. Annu. Rev. 17, 193–284 (1979)
Christodoulou, F. et al. Ancient animal microRNAs and the evolution of tissue identity. Nature 463, 1084–1088 (2010)
Hausdorf, B. et al. Spiralian phylogenomics supports the resurrection of Bryozoa comprising Ectoprocta and Entoprocta. Mol. Biol. Evol. 24, 2723–2729 (2007)
Ebersberger, I., Strauss, S. & von Haeseler, A. HaMStR: profile Hidden Markov Model based search for orthologs in ESTs. BMC Evol. Biol. 9, 157 (2009)
Birney, E., Clamp, M. & Durbin, R. GeneWise and Genomewise. Genome Res. 14, 988–995 (2004)
Katoh, K., Kuma, K.-i., Toh, H. & Miyata, T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 (2005)
Hartmann, S. & Vision, T. Using ESTs for phylogenomics: can one accurately infer a phylogenetic tree from a gappy alignment? BMC Evol. Biol. 8, 95 (2008)
Smith, S. A. & Dunn, C. W. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics 24, 715–716 (2008)
NCBI dbEST (Expressed Sequence Tags Database) 〈http://www.ncbi.nlm.nih.gov/projects/dbEST/〉 (2010)
Helmkampf, M., Bruchhaus, I. & Hausdorf, B. Phylogenomic analyses of lophophorates (brachiopods, phoronids and bryozoans) confirm the Lophotrochozoa concept. Proc. R. Soc. Lond. B 275, 1927–1933 (2008)
Struck, T. H. & Fisse, F. Phylogenetic position of Nemertea derived from phylogenomic data. Mol. Biol. Evol. 25, 728–736 (2008)
Ribosomal Protein Gene Database 〈http://ribosome.med.miyazaki-u.ac.jp〉 (2010)
InParanoid: Eukaryotic Ortholog Groups (100 organisms: 1687023 sequences) 〈http://inparanoid.sbc.su.se/cgi-bin/index.cgi〉 (2010)
Abascal, F., Zardoya, R. & Posada, D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 (2005)
Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006)
Pattengale, N. D., Alipour, M., Bininda-Emonds, O. R. P., Moret, B. M. E. & Stamatakis, A. in RECOMB 2009, LNCS 5541 (ed. Batzoglou, S.) 184–200 (Springer, 2009)
Lartillot, N. & Philippe, H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21, 1095–1109 (2004)
Lartillot, N., Brinkmann, H. & Philippe, H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol. Biol. 7, S4 (2007)
Rambaut, A. & Drummond, A. J. Tracer v1. 4 〈http://beast.bio.ed.ac.uk/Tracer〉 (2007)
Zhou, Y., Rodrigue, N., Lartillot, N. & Philippe, H. Evaluation of the models handling heterotachy in phylogenetic inference. BMC Evol. Biol. 7, 206 (2007)
Zrzavý, J., Riha, P., Pialek, L. & Janouskovec, J. Phylogeny of Annelida (Lophotrochozoa): total-evidence analysis of morphology and six genes. BMC Evol. Biol. 9, 189 (2009)
Rouse, G. W. Trochophore concepts: ciliary bands and the evolution of larvae in spiralian Metazoa. Biol. J. Linn. Soc. 66, 411–464 (1999)
Hartmann-Schröder, G. Teil 58. Annelida, Borstenwürmer, Polychaeta 2nd edn (Gustav Fischer, 1996)
Westheide, W. & Rieger, R. M. Spezielle Zoologie. Erster Teil: Einzeller und Wirbellose Tiere (Gustav Fischer, 1996)
Purschke, G., Arendt, D., Hausen, H. & Müller, M. C. M. Photoreceptor cells and eyes in Annelida. Arthropod Struct. Dev. 35, 211–230 (2006).
Maddison, W. P. & Maddison, D. R. Mesquite: a modular system for evolutionary analysis. Version 2.71. Mesquite Project 〈http://mesquiteproject.org〉 (2009)
Purschke, G., Hessling, R. & Westheide, W. The phylogenetic position of the Clitellata and the Echiura — on the problematic assessment of absent characters. J. Zool. Syst. Evol. Res. 38, 165–173 (2000)
Wiens, J. J. Does adding characters with missing data increase or decrease phylogenetic accuracy? Syst. Biol. 47, 625–640 (1998)
Wiens, J. J., Bonett, R. M. & Chippindale, P. T. Ontogeny discombobulates phylogeny: paedomorphosis and higher-level salamander relationships. Syst. Biol. 54, 91–110 (2005)
Bleidorn, C. The role of character loss in phylogenetic reconstruction as exemplified for the Annelida. J. Zool. Syst. Evol. Res. 45, 299–307 (2007)
Bleidorn, C., Hill, N., Erséus, C. & Tiedemann, R. On the role of character loss in orbiniid phylogeny (Annelida): molecules vs. morphology. Mol. Phylogenet. Evol. 52, 57–69 (2009)
Struck, T. H. Progenetic species in polychaetes (Annelida) and problems assessing their phylogenetic affiliation. Integr. Comp. Biol. 46, 558–568 (2006)
Struck, T. H. Data congruence, paedomorphosis and salamanders. Front. Zool. 4, 22 (2007)
Acknowledgements
We are grateful to I. Ebersberger, S. Strauss and A. von Haeseler for the processing of our raw EST libraries. We also thank M. Aguado for species identification of Typosyllis pigmentata, as well as K. M. Halanych and P. A. Ramey-Balci for suggestions. T.H.S. and C.B. acknowledge support from the marine biological stations in Bamfield, Helgoland, List and Roscoff for collection of annelids. This work was funded by the priority programme ‘Deep Metazoan Phylogeny’ of the Deutsche Forschungsgemeinschaft by grants DFG-STR 683/5-2 (T.H.S.), DFG-Pu-84/5-1 (G.P. and T.H.S.), DFG-TI-349/4-1 (R.T.), DFG Li 998/3-1 (B.L.), DFG-HA 5744/1-1 (S.H.) and DFG-BL-787/2-1 (C.B.).
Author information
Authors and Affiliations
Contributions
T.H.S., G.P., R.T. and C.B. conceived this study. T.H.S. took the lead on data collection of sedentary polychaetes, and writing. T.H.S. and S.H. performed phylogenomic analyses. C.H. aided in the data collection of Sedentaria. C.B. and C.P. took the lead on data collection of errant polychaetes, and C.B., S.H. and N.H. on compilation of the data sets from the EST libraries. A.M. and B.L. generated the EST library of Sipunculus nudus, and M.K. was responsible for the sequencing of the EST libraries. T.H.S., G.P., R.T. and C.B. were the main contributors to the writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Results, additional references, Supplementary Figures 1-8 with legends and Supplementary Tables 1-6. (PDF 1926 kb)
Supplementary Data Set
This file contains the morphological data matrix used in the ancestral reconstructions. Modifications in the character matrix in comparison to Zrzavy et al (2009) are in bold. Converting it to a plain text file allows opening it with Mesquite to show the results of our ancestral reconstructions. (RTF 33 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Struck, T., Paul, C., Hill, N. et al. Phylogenomic analyses unravel annelid evolution. Nature 471, 95–98 (2011). https://doi.org/10.1038/nature09864
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09864
This article is cited by
-
A chromosome-level genome assembly of the Echiura Urechis unicinctus
Scientific Data (2024)
-
Transcriptomic landscape of posterior regeneration in the annelid Platynereis dumerilii
BMC Genomics (2023)
-
The first high-quality chromosome-level genome of the Sipuncula Sipunculus nudus using HiFi and Hi-C data
Scientific Data (2023)
-
CaaX-less lamins: Lophotrochozoa provide a glance at the playground of evolution
Protoplasma (2023)
-
Development and structure of the anterior nervous system and sense organs in the holopelagic annelid Tomopteris spp. (Phyllodocida, Errantia)
Organisms Diversity & Evolution (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.