Abstract
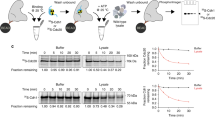
The anaphase-promoting complex or cyclosome (APC/C) is an unusually large E3 ubiquitin ligase responsible for regulating defined cell cycle transitions. Information on how its 13 constituent proteins are assembled, and how they interact with co-activators, substrates and regulatory proteins is limited. Here, we describe a recombinant expression system that allows the reconstitution of holo APC/C and its sub-complexes that, when combined with electron microscopy, mass spectrometry and docking of crystallographic and homology-derived coordinates, provides a precise definition of the organization and structure of all essential APC/C subunits, resulting in a pseudo-atomic model for 70% of the APC/C. A lattice-like appearance of the APC/C is generated by multiple repeat motifs of most APC/C subunits. Three conserved tetratricopeptide repeat (TPR) subunits (Cdc16, Cdc23 and Cdc27) share related superhelical homo-dimeric architectures that assemble to generate a quasi-symmetrical structure. Our structure explains how this TPR sub-complex, together with additional scaffolding subunits (Apc1, Apc4 and Apc5), coordinate the juxtaposition of the catalytic and substrate recognition module (Apc2, Apc11 and Apc10 (also known as Doc1)), and TPR-phosphorylation sites, relative to co-activator, regulatory proteins and substrates.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
EMBL/GenBank/DDBJ
Data deposits
EM maps have been deposited in EMDB with accession numbers EMD-1815 (endogenous cryo-EM APC/CCdh1·D-box), EMD-1816 (endogenous apo APC/C), EMD-1841 (TPR6), EMD-1842 (APC/CΔCde27ΔApc9), EMD-1843 (Apc4–Apc5), EMD-1844 (recombinant apo APC/C), and EMD-1845 (SC8).
References
Peters, J. M. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nature Rev. Mol. Cell Biol. 7, 644–656 (2006)
Thornton, B. R. & Toczyski, D. P. Precise destruction: an emerging picture of the APC. Genes Dev. 20, 3069–3078 (2006)
Sullivan, M. & Morgan, D. O. Finishing mitosis, one step at a time. Nature Rev. Mol. Cell Biol. 8, 894–903 (2007)
Glotzer, M., Murray, A. W. & Kirschner, M. W. Cyclin is degraded by the ubiquitin pathway. Nature 349, 132–138 (1991)
Pfleger, C. M. & Kirschner, M. W. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 14, 655–665 (2000)
Kimata, Y., Baxter, J. E., Fry, A. M. & Yamano, H. A role for the Fizzy/Cdc20 family of proteins in activation of the APC/C distinct from substrate recruitment. Mol. Cell 32, 576–583 (2008)
Eytan, E., Moshe, Y., Braunstein, I. & Hershko, A. Roles of the anaphase-promoting complex/cyclosome and of its activator Cdc20 in functional substrate binding. Proc. Natl Acad. Sci. USA 103, 2081–2086 (2006)
Passmore, L. A. & Barford, D. Coactivator functions in a stoichiometric complex with anaphase-promoting complex/cyclosome to mediate substrate recognition. EMBO Rep. 6, 873–878 (2005)
Sikorski, R. S., Boguski, M. S., Goebl, M. & Hieter, P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 60, 307–317 (1990)
Hirano, T., Kinoshita, N., Morikawa, K. & Yanagida, M. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+ . Cell 60, 319–328 (1990)
Lupas, A., Baumeister, W. & Hofmann, K. A repetitive sequence in subunits of the 26S proteasome and 20S cyclosome (anaphase-promoting complex). Trends Biochem. Sci. 22, 195–196 (1997)
Thornton, B. R. et al. An architectural map of the anaphase-promoting complex. Genes Dev. 20, 449–460 (2006)
Wendt, K. S. et al. Crystal structure of the APC10/DOC1 subunit of the human anaphase-promoting complex. Nature Struct. Biol. 8, 784–788 (2001)
Matyskiela, M. E. & Morgan, D. O. Analysis of activator-binding sites on the APC/C supports a cooperative substrate-binding mechanism. Mol. Cell 34, 68–80 (2009)
Vodermaier, H. C., Gieffers, C., Maurer-Stroh, S., Eisenhaber, F. & Peters, J. M. TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr. Biol. 13, 1459–1468 (2003)
Schwickart, M. et al. Swm1/Apc13 is an evolutionarily conserved subunit of the anaphase-promoting complex stabilizing the association of Cdc16 and Cdc27. Mol. Cell. Biol. 24, 3562–3576 (2004)
Oelschlaegel, T. et al. The yeast APC/C subunit Mnd2 prevents premature sister chromatid separation triggered by the meiosis-specific APC/C-Ama1. Cell 120, 773–788 (2005)
Au, S. W., Leng, X., Harper, J. W. & Barford, D. Implications for the ubiquitination reaction of the anaphase-promoting complex from the crystal structure of the Doc1/Apc10 subunit. J. Mol. Biol. 316, 955–968 (2002)
Zhang, Z. et al. Molecular structure of the N-terminal domain of the APC/C subunit Cdc27 reveals a homo-dimeric tetratricopeptide repeat architecture. J. Mol. Biol. 397, 1316–1328 (2010)
Wang, J., Dye, B. T., Rajashankar, K. R., Kurinov, I. & Schulman, B. A. Insights into anaphase promoting complex TPR subdomain assembly from a CDC26–APC6 structure. Nature Struct. Mol. Biol. 16, 987–989 (2009)
Zhang, Z., Kulkarni, K., Hanrahan, S. J., Thompson, A. J. & Barford, D. The APC/C subunit Cdc16/Cut9 is a contiguous tetratricopeptide repeat superhelix with a homo-dimer interface similar to Cdc27. EMBO J. 29, 3733–3744 (2010)
Han, D. et al. Crystal structure of the N-terminal domain of anaphase-promoting complex subunit 7. J. Biol. Chem. 284, 15137–15146 (2009)
Dube, P. et al. Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. Mol. Cell 20, 867–879 (2005)
Herzog, F. et al. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science 323, 1477–1481 (2009)
Ohi, M. D. et al. Structural organization of the anaphase-promoting complex bound to the mitotic activator Slp1. Mol. Cell 28, 871–885 (2007)
Passmore, L. A. et al. Structural analysis of the anaphase-promoting complex reveals multiple active sites and insights into polyubiquitylation. Mol. Cell 20, 855–866 (2005)
da Fonseca, P. C. et al. Structures of APC/CCdh1 with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature advance online publication. 10.1038/nature09625 (24 November 2010)
Berger, I., Fitzgerald, D. J. & Richmond, T. J. Baculovirus expression system for heterologous multiprotein complexes. Nature Biotechnol. 22, 1583–1587 (2004)
Fitzgerald, D. J. et al. Multiprotein expression strategy for structural biology of eukaryotic complexes. Structure 15, 275–279 (2007)
Passmore, L. A. et al. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. EMBO J. 22, 786–796 (2003)
Passmore, L. A., Barford, D. & Harper, J. W. Purification and assay of the budding yeast anaphase-promoting complex. Methods Enzymol. 398, 195–219 (2005)
Hall, M. C., Torres, M. P., Schroeder, G. K. & Borchers, C. H. Mnd2 and Swm1 are core subunits of the Saccharomyces cerevisiae anaphase-promoting complex. J. Biol. Chem. 278, 16698–16705 (2003)
Kraft, C. et al. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 22, 6598–6609 (2003)
Steen, J. A. et al. Different phosphorylation states of the anaphase promoting complex in response to antimitotic drugs: a quantitative proteomic analysis. Proc. Natl Acad. Sci. USA 105, 6069–6074 (2008)
Sikorski, R. S., Michaud, W. A. & Hieter, P. p62 cdc23 of Saccharomyces cerevisiae: a nuclear tetratricopeptide repeat protein with two mutable domains. Mol. Cell. Biol. 13, 1212–1221 (1993)
Schwab, M., Neutzner, M., Mocker, D. & Seufert, W. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 20, 5165–5175 (2001)
Izawa, D. & Pines, J. Evidence for how APC-C-Cdc20 changes its substrate specificity in mitosis. Nature Cell Biol. doi:10.1038/ncb2165. (in the press)
Hayes, M. J. et al. Early mitotic degradation of Nek2A depends on Cdc20-independent interaction with the APC/C. Nature Cell Biol. 8, 607–614 (2006)
Wolthuis, R. et al. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol. Cell 30, 290–302 (2008)
Duda, D. M. et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134, 995–1006 (2008)
Sobott, F., Hernandez, H., McCammon, M. G., Tito, M. A. & Robinson, C. V. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal. Chem. 74, 1402–1407 (2002)
van Heel, M. et al. Single-particle electron cryo-microscopy: towards atomic resolution. Q. Rev. Biophys. 33, 307–369 (2000)
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996)
Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999)
Gloeckner, C. J., Boldt, K., Schumacher, A., Roepman, R. & Ueffing, M. A novel tandem affinity purification strategy for the efficient isolation and characterisation of native protein complexes. Proteomics 7, 4228–4234 (2007)
Warrens, A. N., Jones, M. D. & Lechler, R. I. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene 186, 29–35 (1997)
Heckman, K. L. & Pease, L. R. Gene splicing and mutagenesis by PCR-driven overlap extension. Nature Protocols 2, 924–932 (2007)
Fitzgerald, D. J. et al. Protein complex expression by using multigene baculoviral vectors. Nature Methods 3, 1021–1032 (2006)
Hernández, H. & Robinson, C. V. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nature Protocols 2, 715–726 (2007)
McKay, A. R., Ruotolo, B. T., Ilag, L. L. & Robinson, C. V. Mass measurements of increased accuracy resolve heterogeneous populations of intact ribosomes. J. Am. Chem. Soc. 128, 11433–11442 (2006)
Taverner, T. et al. Subunit architecture of intact protein complexes from mass spectrometry and homology modeling. Acc. Chem. Res. 41, 617–627 (2008)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Navaza, J., Lepault, J., Rey, F. A., Alvarez-Rua, C. & Borge, J. On the fitting of model electron densities into EM reconstructions: a reciprocal-space formulation. Acta Crystallogr. D 58, 1820–1825 (2002)
Penczek, P. A., Yang, C., Frank, J. & Spahn, C. M. Estimation of variance in single-particle reconstruction using the bootstrap technique. J. Struct. Biol. 154, 168–183 (2006)
Ashkenazy, H., Erez, E., Martz, E., Pupko, T. & Ben-Tal, N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38 (Suppl. 2). W529–W533 (2010)
Landau, M. et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33 (Suppl. 2). W299–W302 (2005)
Acknowledgements
This work was funded by a Cancer Research UK grant to D.B. and EU FP7 PROSPECTS (Proteomics Specification in Space and Time) Grant HEALTH-F4-2008-201648 to C.V.R. We thank F. Beuron for help with electron microscopy and Jing Yang for help with insect cell expression.
Author information
Authors and Affiliations
Contributions
All authors contributed to experimental design, data analysis and manuscript preparation. A.S. generated recombinant APC/C, APC/CΔCdc27ΔApc9, SC8 and Apc4–Apc5 complexes and (with R.I.E.) determined their EM structures. Z.Z. generated recombinant TPR6 and Cdc23–Apc13 and performed MALS. P.C.A.F. and E.H.K. collected and analysed TPR6 EM data and P.C.A.F. determined the TPR6 EM structure. A.S. performed ubiquitylation assays. A.S., R.I.E. and P.C.A.F. fitted coordinates. A.S. and F.S. performed mass spectrometry experiments and A.S., F.S. and C.V.R. analysed mass spectrometry data. E.P.M. helped analyse EM data. D.B. directed the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-20 with legends, Supplementary Tables 1-7 and additional references. (PDF 22487 kb)
Supplementary Movie 1
The movie shows the atomic coordinates of Cdc16-Cdc26, Cdc23, Cdc27, Apc2, (N terminal b-strand of Apc11), Apc10 and Cdh1 were fitted to the 11 Å cryo-EM map of the APC/CCdh1•D-box ternary complex represented in grey mesh. Symmetry related monomers of the Cdc23, Cdc16 and Cdc27 homo-dimers are represented in light and dark orange, red and green, respectively. Other subunits are: Cdh1 (purple), Apc2 (yellow), Apc10 (blue), Cdc26 (cyan), N-terminal b-strand of Apc11 (orange). Red spheres indicate the C-termini of Cdc16, Cdc23 and Apc10, whereas red and blue spheres in Cdc27 denote the N- and C-termini of the inter-TPR insert. The surface molecular boundaries of Apc1 (salmon) and Apc4-Apc5 (green) are indicated. (MOV 30504 kb)
Rights and permissions
About this article
Cite this article
Schreiber, A., Stengel, F., Zhang, Z. et al. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature 470, 227–232 (2011). https://doi.org/10.1038/nature09756
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09756
This article is cited by
-
Nuclear-localized CTEN is a novel transcriptional regulator and promotes cancer cell migration through its downstream target CDC27
Journal of Physiology and Biochemistry (2023)
-
The importance of CDC27 in cancer: molecular pathology and clinical aspects
Cancer Cell International (2021)
-
ERα-related chromothripsis enhances concordant gene transcription on chromosome 17q11.1-q24.1 in luminal breast cancer
BMC Medical Genomics (2020)
-
Mechanisms for the temporal regulation of substrate ubiquitination by the anaphase-promoting complex/cyclosome
Cell Division (2019)
-
Integrated analysis highlights APC11 protein expression as a likely new independent predictive marker for colorectal cancer
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.