Abstract
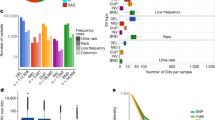
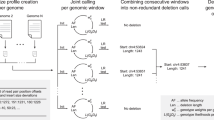
Genomic structural variants (SVs) are abundant in humans, differing from other forms of variation in extent, origin and functional impact. Despite progress in SV characterization, the nucleotide resolution architecture of most SVs remains unknown. We constructed a map of unbalanced SVs (that is, copy number variants) based on whole genome DNA sequencing data from 185 human genomes, integrating evidence from complementary SV discovery approaches with extensive experimental validations. Our map encompassed 22,025 deletions and 6,000 additional SVs, including insertions and tandem duplications. Most SVs (53%) were mapped to nucleotide resolution, which facilitated analysing their origin and functional impact. We examined numerous whole and partial gene deletions with a genotyping approach and observed a depletion of gene disruptions amongst high frequency deletions. Furthermore, we observed differences in the size spectra of SVs originating from distinct formation mechanisms, and constructed a map of SV hotspots formed by common mechanisms. Our analytical framework and SV map serves as a resource for sequencing-based association studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Conrad, D. F. et al. Origins and functional impact of copy number variation in the human genome. Nature 464, 704–712 (2010)
Pinto, D. et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466, 368–372 (2010)
Sebat, J. et al. Strong association of de novo copy number mutations with autism. Science 316, 445–449 (2007)
Stefansson, H. et al. Large recurrent microdeletions associated with schizophrenia. Nature 455, 232–236 (2008)
McCarthy, S. E. et al. Microduplications of 16p11.2 are associated with schizophrenia. Nature Genet. 41, 1223–1227 (2009)
Craddock, N. et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464, 713–720 (2010)
McCarroll, S. A. et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nature Genet. 40, 1107–1112 (2008)
Hastings, P. J., Lupski, J. R., Rosenberg, S. M. & Ira, G. Mechanisms of change in gene copy number. Nature Rev. Genet. 10, 551–564 (2009)
Stankiewicz, P. & Lupski, J. R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 61, 437–455 (2010)
Sebat, J. et al. Large-scale copy number polymorphism in the human genome. Science 305, 525–528 (2004)
Iafrate, A. J. et al. Detection of large-scale variation in the human genome. Nature Genet. 36, 949–951 (2004)
Sharp, A. J. et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 77, 78–88 (2005)
McCarroll, S. A. et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nature Genet. 40, 1166–1174 (2008)
Tuzun, E. et al. Fine-scale structural variation of the human genome. Nature Genet. 37, 727–732 (2005)
Korbel, J. O. et al. Paired-end mapping reveals extensive structural variation in the human genome. Science 318, 420–426 (2007)
Alkan, C. et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nature Genet. 41, 1061–1067 (2009)
Chen, K. et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nature Methods 6, 677–681 (2009)
Hormozdiari, F., Alkan, C., Eichler, E. E. & Sahinalp, S. C. Combinatorial algorithms for structural variation detection in high-throughput sequenced genomes. Genome Res. 19, 1270–1278 (2009)
Medvedev, P., Stanciu, M. & Brudno, M. Computational methods for discovering structural variation with next-generation sequencing. Nature Methods 6, S13–S20 (2009)
McKernan, K. J. et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 19, 1527–1541 (2009)
Chiang, D. Y. et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nature Methods 6, 99–103 (2009)
Kidd, J. M. et al. Mapping and sequencing of structural variation from eight human genomes. Nature 453, 56–64 (2008)
Lee, S., Cheran, E. & Brudno, M. A robust framework for detecting structural variations in a genome. Bioinformatics 24, i59–i67 (2008)
Pang, A. W. et al. Towards a comprehensive structural variation map of an individual human genome. Genome Biol. 11, R52 (2010)
Bailey, J. A. et al. Recent segmental duplications in the human genome. Science 297, 1003–1007 (2002)
Campbell, P. J. et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nature Genet. 40, 722–729 (2008)
Yoon, S., Xuan, Z., Makarov, V., Ye, K. & Sebat, J. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 19, 1586–1592 (2009)
Mills, R. E. et al. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 16, 1182–1190 (2006)
Ye, K., Schulz, M. H., Long, Q., Apweiler, R. & Ning, Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25, 2865–2871 (2009)
Simpson, J. T. et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 19, 1117–1123 (2009)
Hajirasouliha, I. et al. Detection and characterization of novel sequence insertions using paired-end next-generation sequencing. Bioinformatics 26, 1277–1283 (2010)
Li, R. et al. The sequence and de novo assembly of the giant panda genome. Nature 463, 311–317 (2010)
The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010)
Sudmant, P. H. et al. Diversity of human copy number variation and multicopy genes. Science 330, 641–646 (2010)
Willer, C. J. et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nature Genet. 41, 25–34 (2008)
Levy, S. et al. The diploid genome sequence of an individual human. PLoS Biol. 5, e254 (2007)
Hasin-Brumshtein, Y., Lancet, D. & Olender, T. Human olfaction: from genomic variation to phenotypic diversity. Trends Genet. 25, 178–184 (2009)
Hinds, D. A., Kloek, A. P., Jen, M., Chen, X. & Frazer, K. A. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nature Genet. 38, 82–85 (2006)
Altshuler, D. M. et al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010)
Conrad, D. F. et al. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nature Genet. 42, 385–391 (2010)
Lam, H. Y. et al. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nature Biotechnol. 28, 47–55 (2010)
Lupski, J. R. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 14, 417–422 (1998)
Lee, J. A., Carvalho, C. M. & Lupski, J. R. A. DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131, 1235–1247 (2007)
Harrow, J. et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 7, (suppl. 1)S4 (2006)
Acknowledgements
We would like to acknowledge C. Hardy, R. Smith, A. De Witte and S. Giles for their assistance with validation. M.A.B.’s group was supported by a grant from the National Institutes of Health (RO1 GM59290) and G.T.M.’s group by grants R01 HG004719 and RC2 HG005552, also from the NIH. J.O.K.’s group was supported by an Emmy Noether Fellowship of the German Research Foundation (Deutsche Forschungsgemeinschaft). J.W.’s group was supported by the National Basic Research Program of China (973 program no. 2011CB809200), the National Natural Science Foundation of China (30725008; 30890032; 30811130531; 30221004), the Chinese 863 program (2006AA02Z177; 2006AA02Z334; 2006AA02A302; 2009AA022707), the Shenzhen Municipal Government of China (grants JC200903190767A; JC200903190772A; ZYC200903240076A; CXB200903110066A; ZYC200903240077A; ZYC200903240076A and ZYC200903240080A) and the Ole Rømer grant from the Danish Natural Science Research Council. E.E.E.’s group was supported by grants P01 HG004120 and U01 HG005209 from the National Institutes of Health. C.L.’s group was supported by grants from the National Institutes of Health: P41 HG004221, RO1 GM081533 and UO1 HG005209 and X.S. was supported by a T32 fellowship award from the NIH. We thank the Genome Structural Variation Consortium (http://www.sanger.ac.uk/humgen/cnv/42mio/) and the International HapMap Consortium for making available microarray data. The authors acknowledge the individuals participating in the 1000 Genomes Project by providing samples, including the Yoruba people of Ibadan, Nigeria, the community at Beijing Normal University, the people of Tokyo, Japan, and the people of the Utah CEPH community. Furthermore, we thank R. Durbin and L. Steinmetz for comments on the manuscript.
Author information
Authors and Affiliations
Consortia
Contributions
The authors contributed this study at different levels, as described in the following. SV discovery: K.W., C.S., R.E.H., K.C., C.A., A.A., S.C.Y., R.K.C., A.C., Y.F., I.H., F.H., Z.I., D.K., R.Li., Y.L., C.L., R.Lu., X.J.M., H.E.P., L.D., G.T.M., J.S., Ju.W., Ka.Y., Ke.Y., E.E.E., M.B.G., M.E.H., S.A.M. and J.O.K. SV validation: R.E.M., K.W., K.C., A.A., S.C.Y., F.G., M.K.K., J.K., J.N., A.E.U., X.S., A.M.S., J.A.W., Y.Z., Z.D.Z., M.A.B., J.S., M.S., M.E.H., C.L. and J.O.K. SV genotyping: K.W., R.E.H., J.K., J.N., M.E.H. and S.A.M. Data analysis: R.E.M., C.S., C.A., A.A., R.E.H., K.C., S.C.Y., R.K.C., A.C., D.F.C., Y.F., F.H., L.M.I., Z.I., J.M.K., M.K.K., S.K., J.K., E.K., D.K., H.Y.K.L., J.L., R.Li, Y.L., C.L., R.Luo, X.J.M., J.N., H.E.P., T.R., A.S., X.S., M.P.S., J.A.W., Ji.W., Y.Z., Z.D.Z., M.A.B., L.D., G.T.M., G.M., J.S., M.S., Ju.W., Ka.Y., Ke.Y., E.E.E., M.B.G., M.E.H., C.L, S.A.M. and J.O.K. Preparation of manuscript display items: R.E.M., K.W., C.S., C.A., A.A., R.E.H., S.C.Y., L.M.I., S.K., E.K., M.K.K., X.J.M., X.S., J.A.W., M.B.G., S.A.M. and J.O.K. Co-chairs of the Structural Variation Analysis group: E.E.E., M.E.H. and C.L. The following equally contributed to directing the described analyses and participating in the design of the study and should be considered joint senior authors: E.E.E., M.B.G., M.E.H., C.L., S.A.M. and J.O.K. The manuscript was written by the following authors: R.E.M. and J.O.K.
Corresponding author
Ethics declarations
Competing interests
H.E.P. and Y.F. are employees of Life Technologies, the manufacturers of the SOLiD sequencing platform. R.K.C. is an employee of Illumina Cambridge Ltd., the manufacturer of the Illumina sequencing platform.
Additional information
Data sets described here can be obtained from the1000 Genomes Project website at http://www.1000genomes.org (July 2010 Data Release). Individual SV discovery methods can be obtained from sources mentioned in Supplementary Table 2, or upon request from the authors.
Lists of participants and affiliations are shown in Supplementary Information.
Supplementary information
Supplementary Information
This file contains Supplementary Notes, Supplementary Figures 1- 15 with legends, Supplementary Tables 2, 6-8, 12-17, 19 and legends for Supplementary Tables 1-20 (see separate files for Supplementary Tables 1, 3- 5, 9-11, 18 and 20) and Supplementary References. (PDF 3547 kb)
Supplementary Methods
This file contains Supplementary Methods and References. (PDF 281 kb)
Supplementary Table 1
This file contains the sequencing statistics for SV discovery. (XLS 47 kb)
Supplementary Table 3
This file contains a complete list of low coverage calls by institution and set. (ZIP 14400 kb)
Supplementary Table 4
This file contains a complete list of trio calls by institution and set. (ZIP 12314 kb)
Supplementary Table 5
This file contains the Gold standard SV sets for NA12878 and NA12156 from 4 external and orthogonal data sets. (XLS 209 kb)
Supplementary Table 9
This file contains the functional analysis of deletions, which overlap transcripts. (XLS 8530 kb)
Supplementary Table 10
This file contains the Gene Ontology (GO) enrichment analysis for deletions overlapping protein coding regions. (XLS 32 kb)
Supplementary Table 11
This file contains the formation mechanisms and ancestral states of SVs inferred with the BreakSeq pipeline. (XLS 3738 kb)
Supplementary Table 18
This file contains a summary of assembled breakpoints for deletion release set. (XLS 2117 kb)
Supplementary Table 20
This file contains the overlap of partial or whole genotyped, coding region deletions with OMIM Morbid Map. (XLS 26 kb)
Rights and permissions
About this article
Cite this article
Mills, R., Walter, K., Stewart, C. et al. Mapping copy number variation by population-scale genome sequencing. Nature 470, 59–65 (2011). https://doi.org/10.1038/nature09708
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09708
This article is cited by
-
An effect of large-scale deletions and duplications on transcript expression
Functional & Integrative Genomics (2023)
-
CLOVE: classification of genomic fusions into structural variation events
BMC Bioinformatics (2017)
-
Pysim-sv: a package for simulating structural variation data with GC-biases
BMC Bioinformatics (2017)
-
Single-cell template strand sequencing by Strand-seq enables the characterization of individual homologs
Nature Protocols (2017)
-
cnvScan: a CNV screening and annotation tool to improve the clinical utility of computational CNV prediction from exome sequencing data
BMC Genomics (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.