Abstract
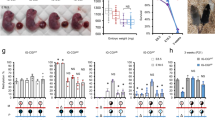
Imprinted genes, defined by their preferential expression of a single parental allele, represent a subset of the mammalian genome and often have key roles in embryonic development1, but also postnatal functions including energy homeostasis2 and behaviour3,4. When the two parental alleles are unequally represented within a social group (when there is sex bias in dispersal and/or variance in reproductive success)5,6, imprinted genes may evolve to modulate social behaviour, although so far no such instance is known. Predominantly expressed from the maternal allele during embryogenesis, Grb10 encodes an intracellular adaptor protein that can interact with several receptor tyrosine kinases and downstream signalling molecules7. Here we demonstrate that within the brain Grb10 is expressed from the paternal allele from fetal life into adulthood and that ablation of this expression engenders increased social dominance specifically among other aspects of social behaviour, a finding supported by the observed increase in allogrooming by paternal Grb10-deficient animals. Grb10 is, therefore, the first example of an imprinted gene that regulates social behaviour. It is also currently alone in exhibiting imprinted expression from each of the parental alleles in a tissue-specific manner, as loss of the peripherally expressed maternal allele leads to significant fetal and placental overgrowth. Thus Grb10 is, so far, a unique imprinted gene, able to influence distinct physiological processes, fetal growth and adult behaviour, owing to actions of the two parental alleles in different tissues.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Reik, W. & Walter, J. Genomic imprinting: parental influence on the genome. Nature Rev. Genet. 2, 21–32 (2001)
Smith, F. M., Garfield, A. S. & Ward, A. Regulation of growth and metabolism by imprinted genes. Cytogenet. Genome Res. 113, 279–291 (2006)
Davies, W., Isles, A. R. & Wilkinson, L. S. Imprinted gene expression in the brain. Neurosci. Biobehav. Rev. 29, 421–430 (2005)
Isles, A. R., Davies, W. & Wilkinson, L. S. Genomic imprinting and the social brain. Phil. Trans. R. Soc. B 361, 2229–2237 (2006)
Haig, D. Genomic imprinting, sex-biased dispersal, and social behavior. Ann. NY Acad. Sci. 907, 149–163 (2000)
Ubeda, F. & Gardner, A. A model for genomic imprinting in the social brain: juveniles. Evolution 64, 2587–2600 (2010)
Holt, L. J. & Siddle, K. Grb10 and Grb14: enigmatic regulators of insulin action—and more? Biochem. J. 388, 393–406 (2005)
Charalambous, M. et al. Disruption of the imprinted Grb10 gene leads to disproportionate overgrowth by an Igf2-independent mechanism. Proc. Natl Acad. Sci. USA 100, 8292–8297 (2003)
Arnaud, P. et al. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum. Mol. Genet. 12, 1005–1019 (2003)
Hikichi, T., Kohda, T., Kaneko-Ishino, T. & Ishino, F. Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes; roles of brain-specific promoters and mouse-specific CTCF-binding sites. Nucleic Acids Res. 31, 1398–1406 (2003)
Monk, D. et al. Reciprocal imprinting of human GRB10 in placental trophoblast and brain: evolutionary conservation of reversed allelic expression. Hum. Mol. Genet. 18, 3066–3074 (2009)
Yamasaki-Ishizaki, Y. et al. Role of DNA methylation and histone H3 lysine 27 methylation in tissue-specific imprinting of mouse Grb10. Mol. Cell. Biol. 27, 732–742 (2007)
Gregg, C. et al. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science 329, 643–648 (2010)
Sanz, L. A. et al. A mono-allelic bivalent chromatin domain controls tissue-specific imprinting at Grb10. EMBO J. 27, 2523–2532 (2008)
Charalambous, M. et al. Maternally-inherited Grb10 reduces placental size and efficiency. Dev. Biol. 337, 1–8 (2010)
Smith, F. M. et al. Mice with a disruption of the imprinted Grb10 gene exhibit altered body composition, glucose homeostasis, and insulin signaling during postnatal life. Mol. Cell. Biol. 27, 5871–5886 (2007)
Plagge, A. et al. Imprinted Nesp55 influences behavioral reactivity to novel environments. Mol. Cell. Biol. 25, 3019–3026 (2005)
Kozlov, S. V. et al. The imprinted gene Magel2 regulates normal circadian output. Nature Genet. 39, 1266–1272 (2007)
Spencer, C. M. et al. Altered anxiety-related and social behaviors in the Fmr1 knockout mouse model of fragile X syndrome. Genes Brain Behav. 4, 420–430 (2005)
Sarna, J. R., Dyck, R. H. & Whishaw, I. Q. The Dalila effect: C57BL6 mice barber whiskers by plucking. Behav. Brain Res. 108, 39–45 (2000)
Keverne, B. Monoallelic gene expression and mammalian evolution. Bioessays 31, 1318–1326 (2009)
Wolf, J. B. & Hager, R. A maternal–offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol. 4, 2238–2243 (2006)
Kaplan, J. R., Manuck, S. B., Fontenot, M. B. & Mann, J. J. Central nervous system monoamine correlates of social dominance in cynomolgus monkeys (Macaca fascicularis). Neuropsychopharmacology 26, 431–443 (2002)
Raleigh, M. J. et al. Serotonergic mechanisms promote dominance acquisition in adult male vervet monkeys. Brain Res. 559, 181–190 (1991)
Edwards, C. A. & Ferguson-Smith, A. C. Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell Biol. 19, 281–289 (2007)
Blagitko, N. et al. Human GRB10 is imprinted and expressed from the paternal and maternal allele in a highly tissue- and isoform-specific fashion. Hum. Mol. Genet. 9, 1587–1595 (2000)
Nagy, A., Gertensenstein, K., Vintersten, K. & Behringer, R. Manipulating the Mouse Embryo: A Laboratory Manual 3rd edn (Cold Spring Harbor Laboratory Press, 2003)
Bennett, W. R., Crew, T. E., Slack, J. M. & Ward, A. Structural-proliferative units and organ growth: effects of insulin-like growth factor 2 on the growth of colon and skin. Development 130, 1079–1088 (2003)
Przydzial, M. J. et al. Nutritional state influences Nociceptin/Orphanin FQ peptide receptor expression in the dorsal raphe nucleus. Behav. Brain Res. 206, 313–317 (2009)
Rousseau, S. J., Jones, I. W., Pullar, I. A. & Wonnacott, S. Presynaptic α7 and non-α7 nicotinic acetylcholine receptors modulate [3H]d-aspartate release from rat frontal cortex in vitro . Neuropharmacology 49, 59–72 (2005)
Ainscough, J. F. et al. Imprinting of Igf2 and H19 from a 130 kb YAC transgene. Development 124, 3621–3632 (1997)
Morales, M. A. et al. Localization of choline acetyltransferase in rat peripheral sympathetic neurons and its coexistence with nitric oxide synthase and neuropeptides. Proc. Natl Acad. Sci. USA 92, 11819–11823 (1995)
Isles, A. R. et al. Urinary odour preferences in mice. Nature 409, 783–784 (2001)
Nevison, C. M. et al. The ownership signature in mouse scent marks is involatile. Proc. R. Soc. Lond. B 270, 1957–1963 (2003)
Doe, C. M. et al. Loss of the imprinted snoRNA mbii-52 leads to increased 5htr2c pre-RNA editing and altered 5HT2CR-mediated behaviour. Hum. Mol. Genet. 18, 2140–2148 (2009)
Dalley, J. W. et al. Distinct changes in cortical acetylcholine and noradrenaline efflux during contingent and noncontingent performance of a visual attentional task. J. Neurosci. 21, 4908–4914 (2001)
Acknowledgements
We thank S. Wonnacott for reagents, the University of Bath Biological Services Unit and S. Routley for technical assistance, I. Jones and P. Mitchell for advice and C. Tickle for comments on the manuscript. We acknowledge funding of the work from the Biotechnology and Biological Sciences Research Council, Medical Research Council, Wellcome Trust and external benefactors.
Author information
Authors and Affiliations
Contributions
A.W. and A.S.G. conceived the project and interpreted the data, with input from L.D.H., A.R.I. and L.S.W.; K.M. and J.E.S.-C. generated the Grb10KO mice; A.S.G. performed most of the experiments with contributions from M.C., J.W.D., S.B., K.G., A.R.I., F.M.S., J.X. and A.W.; A.S.G. and A.W. jointly wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Table 1 and Supplementary Figures 1-6 with legends. (PDF 6698 kb)
Rights and permissions
About this article
Cite this article
Garfield, A., Cowley, M., Smith, F. et al. Distinct physiological and behavioural functions for parental alleles of imprinted Grb10. Nature 469, 534–538 (2011). https://doi.org/10.1038/nature09651
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09651
This article is cited by
-
Advantages of vitrification preservation in assisted reproduction and potential influences on imprinted genes
Clinical Epigenetics (2022)
-
Non-coding de novo mutations in chromatin interactions are implicated in autism spectrum disorder
Molecular Psychiatry (2022)
-
DNA Methylation Analysis of Imprinted Genes in the Cortex and Hippocampus of Cross-Fostered Mice Selectively Bred for Increased Voluntary Wheel-Running
Behavior Genetics (2022)
-
Patients with PWS and related syndromes display differentially methylated regions involved in neurodevelopmental and nutritional trajectory
Clinical Epigenetics (2021)
-
Role of genomic imprinting in mammalian development
Journal of Biosciences (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.