Abstract
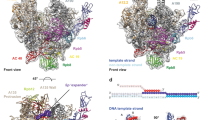
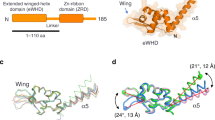
The multi-subunit DNA-dependent RNA polymerase (RNAP) is the principal enzyme of transcription for gene expression. Transcription is regulated by various transcription factors. Gre factor homologue 1 (Gfh1), found in the Thermus genus, is a close homologue of the well-conserved bacterial transcription factor GreA, and inhibits transcription initiation and elongation by binding directly to RNAP1,2,3,4,5,6,7,8. The structural basis of transcription inhibition by Gfh1 has remained elusive, although the crystal structures of RNAP and Gfh1 have been determined separately6,7,8,9. Here we report the crystal structure of Thermus thermophilus RNAP complexed with Gfh1. The amino-terminal coiled-coil domain of Gfh1 fully occludes the channel formed between the two central modules of RNAP; this channel would normally be used for nucleotide triphosphate (NTP) entry into the catalytic site. Furthermore, the tip of the coiled-coil domain occupies the NTP β-γ phosphate-binding site. The NTP-entry channel is expanded, because the central modules are ‘ratcheted’ relative to each other by ∼7°, as compared with the previously reported elongation complexes. This ‘ratcheted state’ is an alternative structural state, defined by a newly acquired contact between the central modules. Therefore, the shape of Gfh1 is appropriate to maintain RNAP in the ratcheted state. Simultaneously, the ratcheting expands the nucleic-acid-binding channel, and kinks the bridge helix, which connects the central modules. Taken together, the present results reveal that Gfh1 inhibits transcription by preventing NTP binding and freezing RNAP in the alternative structural state. The ratcheted state might also be associated with other aspects of transcription, such as RNAP translocation and transcription termination.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Borukhov, S., Polyakov, A., Nikiforov, V. & Goldfarb, A. GreA protein: a transcription elongation factor from Escherichia coli . Proc. Natl Acad. Sci. USA 89, 8899–8902 (1992)
Stebbins, C. E. et al. Crystal structure of the GreA transcript cleavage factor from Escherichia coli . Nature 373, 636–640 (1995)
Vassylyeva, M. N. et al. The carboxy-terminal coiled-coil of the RNA polymerase β'-subunit is the main binding site for Gre factors. EMBO Rep. 8, 1038–1043 (2007)
Hogan, B. P., Hartsch, T. & Erie, D. A. Transcript cleavage by Thermus thermophilus RNA polymerase. Effects of GreA and anti-GreA factors. J. Biol. Chem. 277, 967–975 (2002)
Laptenko, O. & Borukhov, S. Biochemical assays of Gre factors of Thermus thermophilus . Methods Enzymol. 371, 219–232 (2003)
Lamour, V., Hogan, B. P., Erie, D. A. & Darst, S. A. Crystal structure of Thermus aquaticus Gfh1, a Gre-factor paralog that inhibits rather than stimulates transcript cleavage. J. Mol. Biol. 356, 179–188 (2006)
Laptenko, O. et al. pH-dependent conformational switch activates the inhibitor of transcription elongation. EMBO J. 25, 2131–2141 (2006)
Symersky, J. et al. Regulation through the RNA polymerase secondary channel. Structural and functional variability of the coiled-coil transcription factors. J. Biol. Chem. 281, 1309–1312 (2006)
Zhang, G. et al. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell 98, 811–824 (1999)
Cramer, P., Bushnell, D. A. & Kornberg, R. D. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292, 1863–1876 (2001)
Vassylyev, D. G. et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417, 712–719 (2002)
Gnatt, A. L., Cramer, P., Fu, J., Bushnell, D. A. & Kornberg, R. D. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science 292, 1876–1882 (2001)
Vassylyev, D. G., Vassylyeva, M. N., Perederina, A., Tahirov, T. H. & Artsimovitch, I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature 448, 157–162 (2007)
Vassylyev, D. G. et al. Structural basis for substrate loading in bacterial RNA polymerase. Nature 448, 163–168 (2007)
Wang, D., Bushnell, D. A., Westover, K. D., Kaplan, C. D. & Kornberg, R. D. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell 127, 941–954 (2006)
Westover, K. D., Bushnell, D. A. & Kornberg, R. D. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science 303, 1014–1016 (2004)
Westover, K. D., Bushnell, D. A. & Kornberg, R. D. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell 119, 481–489 (2004)
Kettenberger, H., Armache, K. J. & Cramer, P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell 16, 955–965 (2004)
Brueckner, F. & Cramer, P. Structural basis of transcription inhibition by α-amanitin and implications for RNA polymerase II translocation. Nature Struct. Mol. Biol. 15, 811–818 (2008)
Sydow, J. F. et al. Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA. Mol. Cell 34, 710–721 (2009)
Wang, D. et al. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science 324, 1203–1206 (2009)
Temiakov, D. et al. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol. Cell 19, 655–666 (2005)
Tuske, S. et al. Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation. Cell 122, 541–552 (2005)
Tagami, S., Sekine, S., Kumarevel, T., Yamamoto, M. & Yokoyama, S. Crystallization and preliminary X-ray crystallographic analysis of Thermus thermophilus transcription elongation complex bound to Gfh1. Acta Crystallogr. F 66, 64–68 (2010)
Darst, S. A. et al. Conformational flexibility of bacterial RNA polymerase. Proc. Natl Acad. Sci. USA 99, 4296–4301 (2002)
Murakami, K. S., Masuda, S. & Darst, S. A. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 296, 1280–1284 (2002)
Tan, L., Wiesler, S., Trzaska, D., Carney, H. C. & Weinzierl, R. O. Bridge helix and trigger loop perturbations generate superactive RNA polymerases. J. Biol. 7, 40 (2008)
Kettenberger, H., Armache, K. J. & Cramer, P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell 114, 347–357 (2003)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Brunger, A. T. Version 1.2 of the Crystallography and NMR system. Nature Protocols 2, 2728–2733 (2007)
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Brünger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Mukhopadhyay, J. et al. The RNA polymerase “switch region” is a target for inhibitors. Cell 135, 295–307 (2008)
Abyzov, A., Bjornson, R., Felipe, M. & Gerstein, M. RigidFinder: a fast and sensitive method to detect rigid blocks in large macromolecular complexes. Proteins 78, 309–324 (2010)
Vassylyeva, M. N. et al. Purification, crystallization and initial crystallographic analysis of RNA polymerase holoenzyme from Thermus thermophilus . Acta Crystallogr. D 58, 1497–1500 (2002)
Kashkina, E. et al. Elongation complexes of Thermus thermophilus RNA polymerase that possess distinct translocation conformations. Nucleic Acids Res. 34, 4036–4045 (2006)
Chin, J. W., Martin, A. B., King, D. S., Wang, L. & Schultz, P. G. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli . Proc. Natl Acad. Sci. USA 99, 11020–11024 (2002)
Sakamoto, K. et al. Genetic encoding of 3-iodo-L-tyrosine in Escherichia coli for single-wavelength anomalous dispersion phasing in protein crystallography. Structure 17, 335–344 (2009)
Chumpolkulwong, N. et al. Translation of 'rare' codons in a cell-free protein synthesis system from Escherichia coli . J. Struct. Funct. Genomics 7, 31–36 (2006)
Hino, N. et al. Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid. Nature Methods 2, 201–206 (2005)
Acknowledgements
This work is based on experiments performed at SPring-8 (with the approval of the Japan Synchrotron Radiation Research Institute) and at the Swiss Light Source (SLS). We thank N. Shimizu for supporting our data collection at SPring-8 beamline BL41XU; T. Tomizaki and C. Schulze-Briese for supporting our data collection at SLS beamline X06SA; and Y. Fujii for assisting with our data collection and for comments. We thank T. Tanaka and K. Sakamoto for assistance in protein preparation. This work was supported in part by a Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Young Scientists (to S.-i.S.), a JSPS Grant-in-Aid for Scientific Research (to S.i.-S. and S.Y.), and the Targeted Proteins Research Program (TPRP), the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. S.T. was supported by the JSPS Global Centers of Excellence Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms).
Author information
Authors and Affiliations
Contributions
S.T., S.-i.S., T. K. and S.Y. designed the research. S.T. and S.-i.S. performed the structural analysis. M.Y. supported the structural analysis. S.T., S.-i.S., N.H., S.K. and K.S. performed the disulphide-bonding and/or photo-crosslinking analyses. S.T. and Y.M. performed the biochemical analysis of Gre factors. S.-i.S. created the movies. S.T., S.-i.S. and S.Y. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-2, Supplementary Text 1-17, Supplementary Figures 1-27 with legends, Full legends for Supplementary Movies 1-2 and additional references. (PDF 9334 kb)
Supplementary Movie 1
This movie shows the present EC·Gfh1 in the ‘ratcheted’ state and its differences from the ‘tight’ state seen in the previous ECs (see Supplementary information file page 52 for full legend). (MOV 18974 kb)
Supplementary Movie 2
This movie shows the difference in the clamp module orientation between EC·Gfh1 and the previous EC (PDB 2O5I) (see Supplementary information file page 53 for full legend). (MOV 3665 kb)
Rights and permissions
About this article
Cite this article
Tagami, S., Sekine, Si., Kumarevel, T. et al. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature 468, 978–982 (2010). https://doi.org/10.1038/nature09573
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09573
This article is cited by
-
AteqTERT expression and specific tissue activity in a 2-year-old complete plant in Agave tequilana in field conditions
Brazilian Journal of Botany (2020)
-
Interaction effects of DNA, RNA-polymerase, and cellular fluid on the local dynamic behaviors of DNA
Applied Mathematics and Mechanics (2020)
-
Structural basis of RNA polymerase III transcription initiation
Nature (2018)
-
The transcript cleavage factor paralogue TFS4 is a potent RNA polymerase inhibitor
Nature Communications (2017)
-
Structure of a transcribing RNA polymerase II–DSIF complex reveals a multidentate DNA–RNA clamp
Nature Structural & Molecular Biology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.