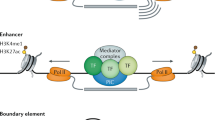
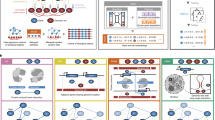
Abstract
Gene regulatory networks (GRNs) provide system level explanations of developmental and physiological functions in the terms of the genomic regulatory code. Depending on their developmental functions, GRNs differ in their degree of hierarchy, and also in the types of modular sub-circuit of which they are composed, although there is a commonly employed sub-circuit repertoire. Mathematical modelling of some types of GRN sub-circuit has deepened biological understanding of the functions they mediate. The structural organization of various kinds of GRN reflects their roles in the life process, and causally illuminates both developmental and evolutionary process.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Oliveri, P., Tu, Q. & Davidson, E. H. Global regulatory logic for specification of an embryonic cell lineage. Proc. Natl Acad. Sci. USA 105, 5955–5962 (2008)This paper provides proof of principle that if a developmental GRN is essentially complete, then it provides causal explanations for the biological functions of the process it controls.
Peter, I. S. & Davidson, E. H. Modularity and design principles in the sea urchin embryo gene regulatory network. FEBS Lett. 583, 3948–3958 (2009)This paper presents the latest comprehensive review of the sea urchin endomesoderm GRN, so far the most extensively validated large scale embryonic GRN, with special emphasis on the topologies of its spatial control sub-circuits.
Davidson, E. H. The Regulatory Genome. Gene Regulatory Networks in Development and Evolution (Academic Press/Elsevier, 2006)
Alon, U. Network motifs: theory and experimental approaches. Nature Rev. Genet. 8, 450–461 (2007)
Mangan, S. & Alon, U. Structure and function of the feed-forward loop network motif. Proc. Natl Acad. Sci. USA 100, 11980–11985 (2003)
Davidson, E. H. Network design principles from the sea urchin embryo. Curr. Opin. Genet. Dev. 19, 535–540 (2009)
Ma, W., Trusina, A., El-Samad, H., Lim, W. A. & Tang, C. Defining network topologies that can achieve biochemical adaptation. Cell 138, 760–773 (2009)
Peter, I. S. & Davidson, E. H. The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stage. Dev. Biol. 340, 188–199 (2010)
Oliveri, P. & Davidson, E. H. Built to run, not fail. Science 315, 1510–1511 (2007)
Koide, T., Hayata, T. & Cho, K. W. Y. Xenopus as a model system to study transcriptional regulatory networks. Proc. Natl Acad. Sci. USA 102, 4943–4948 (2005)
Maduro, M. F. Structure and evolution of the C. elegans embryonic endomesoderm network. Biochim. Biophys. Acta 1789, 250–260 (2009)
Chan, T.-M. et al. Developmental gene regulatory networks in the zebrafish embryo. Biochim. Biophys. Acta 1789, 279–298 (2009)
Morley, R. H. et al. A gene regulatory network directed by zebrafish No tail accounts for its roles in mesoderm formation. Proc. Natl Acad. Sci. USA 106, 3829–3834 (2009)
Stathopoulos, A. & Levine, M. Genomic regulatory networks and animal development. Dev. Cell 9, 449–462 (2005)
Hong, J.-W., Hendrix, D. A., Papatsenko, D. & Levine, M. S. How the Dorsal gradient works: insights from postgenome technologies. Proc. Natl Acad. Sci. USA 105, 20072–20076 (2008)This review summarizes work regulatory control of Dorsal target genes expressed spatially along the dorsal/ventral axis of the syncytial Drosophila embryo.
Liberman, L. M. & Stathopoulos, A. Design flexibility in cis-regulatory control of gene expression: synthetic and comparative evidence. Dev. Biol. 327, 578–589 (2009)This paper presents a novel experimental evidence of cis-regulatory design features in the syncytial dorsal-ventral Drosophila specification system.
Levine, M. & Davidson, E. H. Gene regulatory networks for development. Proc. Natl Acad. Sci. USA 102, 4936–4942 (2005)
Ochoa-Espinosa, A., Yu, D., Tsirigos, A., Struffi, P. & Small, S. Anterior-posterior positional information in the absence of a strong Bicoid gradient. Proc. Natl Acad. Sci. USA 106, 3823–3828 (2009)This paper provides experimental evidence that the anterior/posterior specification system of the Drosophila embryo is controlled by a network of gene interactions rather than only by quantitative positional values of Bicoid.
Liberman, L. M., Teeves, G. T. & Stathopoulos, A. Quantitative imaging of the Dorsal nuclear gradient reveals limitations to threshold-dependent patterning in Drosophila . Proc. Natl Acad. Sci. USA 106, 22317–22322 (2009)
Huang, A. M., Rusch, J. & Levine, M. An anteroposterior Dorsal gradient in the Drosophila embryo. Genes Dev. 11, 1963–1973 (1997)
Saka, Y. & Smith, J. C. A mechanism for sharp transition of morphogen gradient interpretation in Xenopus . BMC Dev. Biol. 7, 47–55 (2007)
Davidson, E. H. Genomic Regulatory Systems: Development and Evolution (Academic Press/Elsevier, 2001)
Su, Y.-H. et al. A perturbation model of the gene regulatory network for oral and aboral ectoderm specification in the sea urchin embryo. Dev. Biol. 329, 410–421 (2009)
Nikitina, N., Sauka-Spengler, T. & Bronner-Fraser, M. Dissecting early regulatory relationships in the lamprey neural crest gene network. Proc. Natl Acad. Sci. USA 105, 20083–20088 (2008)
Woodland, H. R. & Zorn, A. M. The core endodermal gene network of vertebrates: combining developmental precision with evolutionary flexibility. Bioessays 30, 757–765 (2008)
Cvekl, A. & Duncan, M. K. Genetic and epigenetic mechanisms of gene regulation during lens development. Prog. Retin. Eye Res. 26, 555–597 (2007)
Kumar, J. P. The molecular circuitry governing retinal determination. Biochim. Biophys. Acta 1789, 306–314 (2009)
Pimanda, J. E. et al. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc. Natl Acad. Sci. USA 104, 17692–17697 (2007)
Smith, P. A. & Mango, S. E. Role of T-box gene tbx-2 for anterior foregut muscle development in C. elegans . Dev. Biol. 302, 25–39 (2007)
Cripps, R. M. & Olson, E. N. Control of cardiac development by an evolutionarily conserved transcription network. Dev. Biol. 246, 14–28 (2002)
Reim, I., Mohler, J. P. & Frasch, M. Tbx20-related genes, mid and H15 are required for tinman expression, proper patterning, and normal differentiation of cardioblasts in Drosophila . Mech. Dev. 122, 1056–1069 (2005)
Albert, R. & Othmer, H. G. The topology of the regulatory interactions predicts the expression pattern of the segment polarity genes in Drosophila melanogaster . J. Theor. Biol. 223, 1–18 (2003)
Nishi, Y., Ji, H., Wong, W. H., McMahon, A. P. & Vokes, S. A. Modeling the spatio-temporal network that drives patterning in the vertebrate central nervous system. Biochim. Biophys. Acta 1789, 299–305 (2009)
Vokes, S. A., Ji, H., Wong, W. H. & McMahon, A. P. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 22, 2651–2663 (2008)
Ririe, T. O., Fernandes, J. S. & Sternberg, P. W. The Caenorhabditis elegans vulva: A post-embryonic gene regulatory network controlling organogenesis. Proc. Natl Acad. Sci. USA 105, 20095–20099 (2008)
Graf, T. & Enver, T. Forcing cells to change lineages. Nature 462, 587–594 (2009)This review comprehensively traverses the process of terminal lineage fate choice in pluripotential hematopoietic systems.
Swiers, G., Patient, R. & Loose, M. Genetic regulatory networks programming hematopoietic stem cells and erythroid lineage specification. Dev. Biol. 294, 525–540 (2006)
Laslo, P. et al. Multilineage transcription priming and determination of alternate hematopoietic cell fates. Cell 126, 755–766 (2006)This paper exemplifies a commonly used mathematical approach invoking bi-stable state kinetics to explain lineage choice.
Smith, J. & Davidson, E. H. Gene regulatory network subcircuit controlling a dynamic spatial pattern of signaling in the sea urchin embryo. Proc. Natl Acad. Sci. USA 105, 20089–20094 (2008)
Zhang, P. et al. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc. Natl Acad. Sci. USA 96, 8705–8710 (1999)
Huang, S., Guo, Y.-P., May, G. & Enver, T. Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev. Biol. 305, 695–713 (2007)
Stopka, T., Amanatullah, D. F., Papetti, M. & Skoultchi, A. I. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J. 24, 3712–3723 (2005)
Starck, J. et al. Functional cross-antagonism between transcription factors FLI-1 and EKLF. Mol. Cell. Biol. 23, 1390–1402 (2003)
Rothenberg, E. V. Decision by committee: new light on the CD4/CD8-lineage choice. Immunol. Cell Biol. 87, 109–112 (2009)
Wang, L. & Bosselut, R. CD4–CD8 lineage differentiation: Thpok-ing into the nucleus. J. Immunol. 183, 2903–2910 (2009)
Setoguchi, R. et al. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science 319, 822–825 (2008)
Narula, J., Smith, A. M. & Gottgens, B. and Igoshin, O. A. Modeling reveals bistability and low-pass filtering in the network module determining blood stem cell fate. PLoS Comput. Biol. 6, e1000771 (2010)
Hu, M. et al. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 11, 774–785 (1997)
Miyamoto, T. et al. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell 3, 137–147 (2002)
Lagha, M. et al. Pax3:Foxc2 reciprocal repression in the somite modulates muscular versus vascular cell fate choice in multipotent progenitors. Dev. Cell 17, 892–899 (2009)
Johnson, R. J., Jr, Chang, S., Etchberger, J. F., Ortiz, C. O. & Hobert, O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl Acad. Sci. USA 102, 12449–12454 (2005)
Vierbuchen, T. et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041 (2010)
Zhou, Q., Brown, J., Kanarek, A., Rajagopal, J. & Melton, D. A. In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nature 455, 627–632 (2008)
Gilchrist, M. et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature 441, 173–178 (2006)
Hobert, O. Regulatory logic of neuronal diversity: Terminal selector genes and selector motifs. Proc. Natl Acad. Sci. USA 105, 20067–20071 (2008)
Bröhl, D. et al. A transcriptional network coordinately determines transmitter and peptidergic fate in the dorsal spinal chord. Dev. Biol. 322, 381–393 (2008)
Yun, K. & Wold, B. Skeletal muscle determination and differentiation: story of a core regulatory network and its context. Curr. Opin. Cell Biol. 8, 877–889 (1996)
Pan, G. & Thomson, J. A. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 17, 42–49 (2007)
Boyer, L. A. et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 (2005)
Zhou, Q., Chipperfield, H., Melton, D. A. & Wong, W. H. A gene regulatory network in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA 104, 16438–16443 (2007)
Boyer, L. A. et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 (2006)
Mortazavi, A., Chen Leeper Thompson, E., Garcia, S. T., Myers, R. M. & Wold, B. Comparative genomics modeling of the NRSF/REST repressor network: from single conserved sites to genome-wide repertoire. Genome Res. 16, 1208–1221 (2006)
Zhu, X. & Rosenfeld, M. G. Transcriptional control of precursor proliferation in the early phases of pituitary development. Curr. Opin. Genet. Dev. 14, 567–574 (2004)
Bessa, J. et al. meis1 regulates cyclin D1 and c-myc expression, and controls the proliferation of the multipotent cells in the early developing zebrafish eye. Development 135, 799–803 (2008)
Christiaen, L. et al. The transcription/migration interface in heart precursors of Ciona intestinalis . Science 320, 1349–1352 (2008)This paper presents direct evidence of the regulatory structure of a morphogenetic gene cassette, showing that only certain key genes are controlled by the upstream GRN while a majority are expressed anyway.
Chanut-Delalande, H., Fernandes, I., Roch, F., Payre, F. & Plaza, S. Shavenbaby couples patterning to epidermal cell shape control. PLoS Biol. 4, 1549–1561 (2006)
Amit, I. et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326, 257–263 (2009)This paper presents the most complete analysis yet available of structure and function in a physiological GRN.
Rosenfeld, N. & Alon, U. Response delays and the structure of transcription networks. J. Mol. Biol. 329, 645–654 (2003)
Bolouri, H. Computational Modeling of Gene Regulatory Networks – A Primer (Imperial College Press, 2008)
Sánchez, L. & Thieffry, D. A logical analysis of the gap gene system. J. Theor. Biol. 211, 115–141 (2001)
Jaeger, J. et al. Dynamical analysis of regulatory interactions in the gap gene system of Drosophila melanogaster . Genetics 167, 1721–1737 (2004)
Perkins, T. J., Jaeger, J., Reintz, J. & Glass, L. Reverse engineering the gap gene network of Drosophila melanogaster . PLoS Comput. Biol. 2, e051 (2006)This paper provides a comprehensive computational treatment of the Drosophila gap gene network using estimates of numerous constants obtained by high resolution imaging.
Rivera-Pomar, R. & Jaeckle, H. From gradients to stripes in Drosophila mebryogenesis: filling in the gaps. Trends Genet. 12, 478–483 (1996)
Kraut, R. & Levine, M. Mutually repressive interactions between the gap genes giant and Krüpple define middle body regions of the Drosophila embryo. Development 111, 611–621 (1991)
Segal, E., Raveh-Sadka, T., Schroeder, M., Unnerstall, U. & Gaul, U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature 451, 535–540 (2008)
Lembong, J., Yakoby, N. & Shvartsman, S. Y. Pattern formation by dynamically interacting network motifs. Proc. Natl Acad. Sci. USA 106, 3213–3218 (2009)
Dessaud, E. et al. Dynamic assignment and maintenance of positional identity in the ventral neural tube by the morphogen Sonic hedgehog. PLoS Biol. 8, e1000382 (2010)This paper provides a new insight into how positional values of Hedgehog ligand are used to set transcriptional thresholds.
Ribes, V. & Briscoe, J. Establishing and interpreting graded Sonic Hedgehog signaling during vertebrate neural tube patterning: the role of negative feedback. Cold Spring Harb. Perspect. Biol. 1, a002014 (2009)
Goentoro, L., Shoval, O., Kirschner, M. W. & Alon, U. The incoherent feedforward loop can provide fold-change detection in gene regulation. Mol. Cell 36, 894–899 (2009)This analysis shows how a common GRN sub-circuit can operate to interpret relative changes in signal strength.
Goentoro, L. & Kirschner, M. W. Evidence that fold-change, and not absolute level, of β-catenin dictates Wnt signaling. Mol. Cell 36, 872–884 (2009)
Spooner, C. J. et al. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity 31, 576–586 (2009)
Chickarmane, V., Enver, T. & Peterson, C. Computational modeling of the hematopoietic erythroid-myeloid switch reveals insights into cooperativity, priming, and irreversibility. PLoS Comput. Biol. 5, e1000268 (2009)This paper presents an alternative computational treatment of lineage choice in a haematopoietic system.
Shea, M. A. & Ackers, G. K. The OR control system of bacteriophage lambda: A physical-chemical model for gene regulation. J. Mol. Biol. 181, 211–230 (1985)
Bolouri, H. & Davidson, E. H. Transcriptional regulatory cascades in development: Initial rates, not steady state, determine network kinetics. Proc. Natl Acad. Sci. USA 100, 9371–9376 (2003)This paper models sea urchin regulatory cascade kinetics and demonstrates using measured constants that genes are successively activated long before any of the transcriptional functions attain steady state.
Materna, S. C., Nam, J. & Davidson, E. H. High accuracy, high-resolution prevalence measurement for the majority of locally expressed regulatory genes in early sea urchin development. Gene Expr. Patterns 10, 177–184 (2010)
Davidson, E. H. & Erwin, D. H. Gene regulatory networks and the evolution of animal body plans. Science 311, 796–800 (2006)This paper introduced the theory that highly conserved GRN sub-circuits account for the phylogenetic distribution of major characters of the animal body plan.
Erwin, D. H. & Davidson, E. H. The evolution of hierarchical gene regulatory networks. Nature Rev. Genet. 10, 141–148 (2009)
Davidson, E. H. & Erwin, D. H. An integrated view of Precambrian eumetazoan evolution. Cold Spring Harb. Symp. Quant. Biol. 74, 65–80 (2009)
Gao, F. & Davidson, E. H. Transfer of a large gene regulatory apparatus to a new developmental address in echinoid evolution. Proc. Natl Acad. Sci. USA 105, 6091–6096 (2008)
Hinman, V. F. & Davidson, E. H. Evolutionary plasticity of developmental gene regulatory network architecture. Proc. Natl Acad. Sci. USA 104, 19404–19409 (2007)
Hinman, V. F., Yankura, K. A. & McCauley, B. S. Evolution of gene regulatory network architectures: Examples of subcircuit conservation and plasticity between classes of echinoderms. Biochim. Biophys. Acta 1789, 326–332 (2009)
Bolouri, H. & Davidson, E. H. The gene regulatory network basis of the “community effect,” and analysis of a sea urchin embryo example. Dev. Biol. 340, 170–178 (2010)
Acknowledgements
I am grateful for the reviews of the manuscript by E. V. Rothenberg and I. S. Peter. This work was supported by NIH grants HD-37105 and GM-61005 and by the Lucille P. Markey Charitable Trust.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
PowerPoint slides
Rights and permissions
About this article
Cite this article
Davidson, E. Emerging properties of animal gene regulatory networks. Nature 468, 911–920 (2010). https://doi.org/10.1038/nature09645
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09645
This article is cited by
-
Plasticity-led evolution as an intrinsic property of developmental gene regulatory networks
Scientific Reports (2023)
-
The evolution of complex multicellularity in animals
Biology & Philosophy (2022)
-
A mechanistic framework for cardiometabolic and coronary artery diseases
Nature Cardiovascular Research (2022)
-
Three topological features of regulatory networks control life-essential and specialized subsystems
Scientific Reports (2021)
-
Riding the crest to get a head: neural crest evolution in vertebrates
Nature Reviews Neuroscience (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.