Abstract
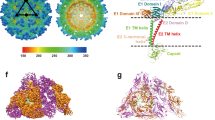
Chikungunya virus (CHIKV) is an emerging mosquito-borne alphavirus that has caused widespread outbreaks of debilitating human disease in the past five years1. CHIKV invasion of susceptible cells is mediated by two viral glycoproteins, E1 and E2, which carry the main antigenic determinants and form an icosahedral shell at the virion surface. Glycoprotein E2, derived from furin cleavage of the p62 precursor into E3 and E2, is responsible for receptor binding, and E1 for membrane fusion. In the context of a concerted multidisciplinary effort to understand the biology of CHIKV2, here we report the crystal structures of the precursor p62–E1 heterodimer and of the mature E3–E2–E1 glycoprotein complexes. The resulting atomic models allow the synthesis of a wealth of genetic, biochemical, immunological and electron microscopy data accumulated over the years on alphaviruses in general. This combination yields a detailed picture of the functional architecture of the 25 MDa alphavirus surface glycoprotein shell. Together with the accompanying report on the structure of the Sindbis virus E2–E1 heterodimer at acidic pH (ref. 3), this work also provides new insight into the acid-triggered conformational change on the virus particle and its inbuilt inhibition mechanism in the immature complex.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Protein Data Bank
Data deposits
Structure factors and coordinates for the structures of the p62–E1 (crystal IO), E3–E2–E1sp (crystal MM), E3–E2–E1f (crystal MO1), E3–E2–E1t (crystal MO2) and Os2-E3–E2–E1t (crystal MO3) complexes have been deposited in the Protein Data Bank under accession codes 3N40, 3N41, 3N42, 3N43 and 3N44, respectively. The coordinates of the molecules fitted in the cryo-EM 3D reconstructions of SINV and SFV particles were deposited under accession codes 2XFB and 2XFC, respectively.
References
Her, Z., Kam, Y. W., Lin, R. T. & Ng, L. F. Chikungunya: a bending reality. Microbes Infect. 11, 1165–1176 (2009)
Schwartz, O. & Albert, M. L. Biology and pathogenesis of chikungunya virus. Nature Rev. Microbiol. 8, 491–500 (2010)
Li, L., Jose, J., Xiang, Y., Kuhn, R. J. & Rossmann, M. G. Structural changes of envelope proteins during alphavirus fusion. Nature 10.1038/nature09546 (this issue)
Halstead, S. B. in Pediatric Infectious Diseases (eds Feigin, R. & Cherry, J.) 2178–2183 (Saunders, 2004)
Robinson, M. C. An epidemic of virus disease in southern province, Tanganyika territory, in 1952–1953 I. Clinical features. Trans. R. Soc. Trop. Med. Hyg. 49, 28–32 (1955)
Johnson, F. Notes on Kimakonde. Bull. Sch. Orient. Studies 2, 417–466 (1922)
Carey, D. E. Chikungunya and dengue: a case of mistaken identity? J. Hist. Med. Allied Sci. 26, 243–262 (1971)
Schuffenecker, I. et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 3, e263 (2006)
Tsetsarkin, K. A., Vanlandingham, D. L., McGee, C. E. & Higgs, S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 3, e201 (2007)
Vazeille, M. et al. Two chikungunya isolates from the outbreak of La Reunion (Indian Ocean) exhibit different patterns of infection in the mosquito, aedes albopictus . PLoS ONE 2, e1168 (2007)
Enserink, M. Entomology: a mosquito goes global. Science 320, 864–866 (2008)
Akahata, W. et al. A virus-like particle vaccine for epidemic chikungunya virus protects nonhuman primates against infection. Nature Med. 16, 334–338 (2010)
Salminen, A. et al. Membrane fusion process of Semliki Forest virus. II. Cleavage-dependent reorganization of the spike protein complex controls virus entry. J. Cell Biol. 116, 349–357 (1992)
Ziemiecki, A., Garoff, H. & Simons, K. Formation of the Semliki Forest virus membrane glycoprotein complexes in the infected cell. J. Gen. Virol. 50, 111–123 (1980)
Marsh, M. & Helenius, A. Virus entry into animal cells. Adv. Virus Res. 36, 107–151 (1989)
Wahlberg, J. M., Boere, W. A. & Garoff, H. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J. Virol. 63, 4991–4997 (1989)
Kielian, M. & Helenius, A. pH-induced alterations in the fusogenic spike protein of Semliki Forest virus. J. Cell Biol. 101, 2284–2291 (1985)
Wahlberg, J. M., Bron, R., Wilschut, J. & Garoff, H. Membrane fusion of Semliki Forest virus involves homotrimers of the fusion protein. J. Virol. 66, 7309–7318 (1992)
Lescar, J. et al. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105, 137–148 (2001)
Gibbons, D. L. et al. Conformational change and protein–protein interactions of the fusion protein of Semliki Forest virus. Nature 427, 320–325 (2004)
Kielian, M. & Rey, F. A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nature Rev. Microbiol. 4, 67–76 (2006)
Rey, F. A. et al. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375, 291–298 (1995)
Lobigs, M., Zhao, H. X. & Garoff, H. Function of Semliki Forest virus E3 peptide in virus assembly: replacement of E3 with an artificial signal peptide abolishes spike heterodimerization and surface expression of E1. J. Virol. 64, 4346–4355 (1990)
Tsetsarkin, K. A. et al. Epistatic roles of E2 glycoprotein mutations in adaption of chikungunya virus to Aedes albopictus and Ae. aegypti mosquitoes. PLoS ONE 4, e6835 (2009)
Pierro, D. J., Powers, E. L. & Olson, K. E. Genetic determinants of Sindbis virus mosquito infection are associated with a highly conserved alphavirus and flavivirus envelope sequence. J. Virol. 82, 2966–2974 (2008)
Meyer, W. J. & Johnston, R. E. Structural rearrangement of infecting Sindbis virions at the cell surface: mapping of newly accessible epitopes. J. Virol. 67, 5117–5125 (1993)
Meyer, W. J. et al. Conformational alteration of Sindbis virion glycoproteins induced by heat, reducing agents, or low pH. J. Virol. 66, 3504–3513 (1992)
Flynn, D. C., Meyer, W. J., Mackenzie, J. M., Jr & Johnston, R. E. A conformational change in Sindbis virus glycoproteins E1 and E2 is detected at the plasma membrane as a consequence of early virus-cell interaction. J. Virol. 64, 3643–3653 (1990)
Wu, S. R. et al. The dynamic envelope of a fusion class II virus. Prefusion stages of Semliki Forest virus revealed by electron cryomicroscopy. J. Biol. Chem. 282, 6752–6762 (2007)
Mukhopadhyay, S. et al. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 14, 63–73 (2006)
Schuffenecker, I. et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 3, e263 (2006)
Krey, T. et al. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 6, e1000762 (2010)
Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D 66, 133–144 (2010)
Evans, P. R. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2005)
Collaborative Computational Project Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Navaza, J. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D 57, 1367–1372 (2001)
Roussel, A. et al. Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure 14, 75–86 (2006)
Bricogne, G. et al. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D 59, 2023–2030 (2003)
Abrahams, J. P. & Leslie, A. G. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D 52, 30–42 (1996)
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D 62, 1002–1011 (2006)
Cowtan, K. ‘dm’: an automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newsl. Protein Crystallogr. 31, 34–38 (1994)
Emsley, P. et al. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Bricogne, G. et al. BUSTER version 2.9. (Global Phasing, 2010)
Painter, J. & Merritt, E. A. A molecular viewer for the analysis of TLS rigid-body motion in macromolecules. Acta Crystallogr. D 61, 465–471 (2005)
Mukhopadhyay, S. et al. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 14, 63–73 (2006)
Mancini, E. J. et al. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol. Cell 5, 255–266 (2000)
Akahata, W. et al. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nature Med. 16, 334–338 (2010)
Navaza, J. et al. On the fitting of model electron densities into EM reconstructions: a reciprocal-space formulation. Acta Crystallogr. D 58, 1820–1825 (2002)
Acknowledgements
We thank the CHIKV task force at Institut Pasteur, in particular the group of F. Tangy and the staff of platform PF8 for the CHIKV complementary DNA; A. Haouz of PF6 for crystallogenesis; the staff of synchrotron beamlines PROXIMA 1 at Soleil, ID23-eh2 at the European Synchrotron Radiation Facility and PX-I at the Swiss Light Source; M. Rossmann and Y. Sun for providing the 16 Å cryo-EM map of CHIKV virion-like particles and for sharing the coordinates and manuscript of the low pH structure of the SINV E1–E2 heterodimer before publication; and members of the F.A.R. laboratory for help during data collection. J.E.V. was supported by a Marie Curie fellowship through the European Union Research Traning Network program “Intrapath”. This work was funded in part by the French ‘Agence Nationale de la Recherche’ grant DENtry in the program ‘Microbiologie, Infections et Immunité’, by Merck-Serono, by the Pediatric Dengue Vaccine Initiative and by the Institut Pasteur program PTR201 CHIKV to F.A.R..
Author information
Authors and Affiliations
Contributions
J.E.V. made the constructs, produced and purified the protein, grew the crystals and participated in diffraction data collection; analysed the literature and prepared tables and figures. M.-C.V. carried out most of the various crystallographic refinements and prepared the figures, S.D. carried out the fitting into the cryo-EM maps of various alphavirus particles and prepared the figures, C.G.-B. and E.C. participated in optimizing protein production in large scale for crystal trials, C.V. and G.B. participated in data processing and in the structure determination. A.T. carried out specific data collection strategies to improve the signal to noise to extract anomalous signal for phasing; F.A.R. conceived the experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Analysis, Supplementary References, Supplementary Tables 1-7 and Supplementary Figures 1-7 with legends. (ZIP 12830 kb)
Supplementary Movie 1
This movie shows Six spikes around a Q6 axis of the T=4 icosahedral lattice (see legend in Supplementary Information). (MOV 4582 kb)
Rights and permissions
About this article
Cite this article
Voss, J., Vaney, MC., Duquerroy, S. et al. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468, 709–712 (2010). https://doi.org/10.1038/nature09555
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09555
This article is cited by
-
LDLR is used as a cell entry receptor by multiple alphaviruses
Nature Communications (2024)
-
Peptide aptamer-based time-resolved fluoroimmunoassay for CHIKV diagnosis
Virology Journal (2023)
-
An overview of the role of Niemann-pick C1 (NPC1) in viral infections and inhibition of viral infections through NPC1 inhibitor
Cell Communication and Signaling (2023)
-
Chikungunya fever
Nature Reviews Disease Primers (2023)
-
Chikungunya virus cell-to-cell transmission is mediated by intercellular extensions in vitro and in vivo
Nature Microbiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.