Abstract
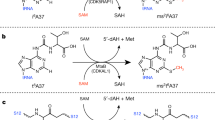
Archaeal and eukaryotic translation elongation factor 2 contain a unique post-translationally modified histidine residue called diphthamide, which is the target of diphtheria toxin. The biosynthesis of diphthamide was proposed to involve three steps, with the first being the formation of a C–C bond between the histidine residue and the 3-amino-3-carboxypropyl group of S-adenosyl-l-methionine (SAM). However, further details of the biosynthesis remain unknown. Here we present structural and biochemical evidence showing that the first step of diphthamide biosynthesis in the archaeon Pyrococcus horikoshii uses a novel iron–sulphur-cluster enzyme, Dph2. Dph2 is a homodimer and each of its monomers can bind a [4Fe–4S] cluster. Biochemical data suggest that unlike the enzymes in the radical SAM superfamily, Dph2 does not form the canonical 5′-deoxyadenosyl radical. Instead, it breaks the Cγ,Met–S bond of SAM and generates a 3-amino-3-carboxypropyl radical. Our results suggest that P. horikoshii Dph2 represents a previously unknown, SAM-dependent, [4Fe–4S]-containing enzyme that catalyses unprecedented chemistry.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Centers for Disease Control and Prevention. Diphtheria. Disease Listing, Diphtheria, Technical Information | CDC Bacterial, Mycotic Diseases 〈http://www.cdc.gov/ncidod/DBMD/diseaseinfo/diptheria_t.htm〉 (2005)
Collier, R. J. Understanding the mode of action of diphtheria toxin: a perspective on progress during the 20th century. Toxicon 39, 1793–1803 (2001)
Liu, S., Milne, G. T., Kuremsky, J. G., Fink, G. R. & Leppla, S. H. Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol. Cell. Biol. 24, 9487–9497 (2004)
Gomez-Lorenzo, M. G. et al. Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 Å resolution. EMBO J. 19, 2710–2718 (2000)
Ortiz, P. A., Ulloque, R., Kihara, G. K., Zheng, H. & Kinzy, T. G. Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J. Biol. Chem. 281, 32639–32648 (2006)
Walsh, C. T. Posttranslational Modifications of Proteins: Expanding Nature’s Inventory 321–323 (Roberts, 2006)
Moehring, J. M., Moehring, T. J. & Danley, D. E. Posttranslational modification of elongation factor 2 in diphtheriatoxin-resistant mutants of CHO-K1 cells. Proc. Natl Acad. Sci. USA 77, 1010–1014 (1980)
Moehring, T. J., Danley, D. E. & Moehring, J. M. In vitro biosynthesis of diphthamide, studied with mutant Chinese hamster ovary cells resistant to diphtheria toxin. Mol. Cell. Biol. 4, 642–650 (1984)
Chen, J. Y., Bodley, J. W. & Livingston, D. M. Diphtheria toxin-resistant mutants of Saccharomyces cerevisiae . Mol. Cell. Biol. 5, 3357–3360 (1985)
Mattheakis, L. C., Shen, W. H. & Collier, R. J. DPH5, a methyltransferase gene required for diphthamide biosynthesis in Saccharomyces cerevisiae . Mol. Cell. Biol. 12, 4026–4037 (1992)
Mattheakis, L. C., Sor, F. & Collier, R. J. Diphthamide synthesis in Saccharomyces cerevisiae: structure of the DPH2 gene. Gene 132, 149–154 (1993)
Phillips, N. J., Ziegler, M. R. & Deaven, L. L. A cDNA from the ovarian cancer critical region of deletion on chromosome 17p13.3. Cancer Lett. 102, 85–90 (1996)
Schultz, D. C., Balasara, B. R., Testa, J. R. & Godwin, A. K. Cloning and localization of a human diphthamide biosynthesis-like protein-2 gene, DPH2L2 . Genomics 52, 186–191 (1998)
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990)
Frey, P. A., Hegeman, A. D. & Ruzicka, F. J. The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 43, 63–88 (2008)
Sakuraba, H., Tsuge, H., Yoneda, K., Katunuma, N. & Ohshima, T. Crystal structure of the NAD biosynthetic enzyme quinolinate synthase. J. Biol. Chem. 280, 26645–26648 (2005)
Sofia, H. J., Chen, G., Hetzler, B. G., Reyes-Spindola, J. F. & Miller, N. E. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29, 1097–1106 (2001)
Chatterjee, A. et al. Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily. Nature Chem. Biol. 4, 758–765 (2008)
McGlynn, S. E. et al. Identification and characterization of a novel member of the radical AdoMet enzyme superfamily and implications for the biosynthesis of the Hmd hydrogenase active site cofactor. J. Bacteriol. 192, 595–598 (2010)
Makinen, G. B. & Wells, M. W. in ENDOR, EPR and Electron Spin Echo for Probing Coordination Spheres (eds Sigel, H. & Sigel, A.) 129–204 (Dekker, 1987)
Lieder, K. W. et al. S-adenosylmethionine-dependent reduction of lysine 2,3-aminomutase and observation of the catalytically functional iron–sulfur centers by electron paramagnetic resonance. Biochemistry 37, 2578–2585 (1998)
Walsby, C. J. et al. Electron-nuclear double resonance spectroscopic evidence that S-adenosylmethionine binds in contact with the catalytically active [4Fe–4S]+ cluster of pyruvate formate-lyase activating enzyme. J. Am. Chem. Soc. 124, 3143–3151 (2002)
Cicchillo, R. M. et al. Escherichia coli quinolinate synthetase does indeed harbor a [4Fe-4S] cluster. J. Am. Chem. Soc. 127, 7310–7311 (2005)
Saunders, A. H. et al. Characterization of quinolinate synthases from Escherichia coli, Mycobacterium tuberculosis, and Pyrococcus horikoshii indicates that [4Fe-4S] clusters are common cofactors throughout this class of enzymes. Biochemistry 47, 10999–11012 (2008)
Berkovitch, F., Nicolet, Y., Wan, J. T., Jarrett, J. T. & Drennan, C. L. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science 303, 76–79 (2004)
Layer, G., Moser, J., Heinz, D. W., Jahn, D. & Schubert, W.-D. Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of radical SAM enzymes. EMBO J. 22, 6214–6224 (2003)
Lepore, B. W., Ruzicka, F. J., Frey, P. A. & Ringe, D. The X-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale . Proc. Natl Acad. Sci. USA 102, 13819–13824 (2005)
Hänzelmann, P. & Schindelin, H. Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc. Natl Acad. Sci. USA 101, 12870–12875 (2004)
Vey, J. L. et al. Structural basis for glycyl radical formation by pyruvate formate-lyase activating enzyme. Proc. Natl Acad. Sci. USA 105, 16137–16141 (2008)
Marinoni, I. et al. Characterization of l-aspartate oxidase and quinolinate synthase from Bacillus subtilis . FEBS J. 275, 5090–5107 (2008)
Rekittke, I. et al. Structure of (E)-4-hydroxy-3-methyl-but-2-enyl diphosphate reductase, the terminal enzyme of the non-mevalonate pathway. J. Am. Chem. Soc. 130, 17206–17207 (2008)
Walsby, C. J., Ortillo, D., Broderick, W. E., Broderick, J. B. & Hoffman, B. M. An anchoring role for FeS clusters: chelation of the amino acid moiety of S-adenosylmethionine to the unique iron site of the [4Fe–4S] cluster of pyruvate formate-lyase activating enzyme. J. Am. Chem. Soc. 124, 11270–11271 (2002)
Krogan, N. J. et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae . Nature 440, 637–643 (2006)
Gavin, A.-C. et al. Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 (2006)
Collins, S. R. et al. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae . Mol. Cell. Proteomics 6, 439–450 (2007)
Proudfoot, M. et al. Biochemical and structural characterization of a novel family of cystathionine β-synthase domain proteins fused to a Zn ribbon-like domain. J. Mol. Biol. 375, 301–315 (2008)
Johnson, D. C., Dean, D. R., Smith, A. D. & Johnson, M. K. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281 (2005)
Lill, R. & Mühlenhoff, U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 77, 669–700 (2008)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008)
Collaborative. Computation Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Terwilliger, T. C. Reciprocal-space solvent flattening. Acta Crystallogr. D 55, 1863–1871 (1999)
Terwilliger, T. C. Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972 (2000)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Brünger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Acknowledgements
We thank L. Kinsland for assistance with manuscript preparation, the Dreyfus Foundation for a New Faculty Award to H.L., NIH/NIGMS R01GM088276 to H.L. and S.E.E., and NIH/NCRR P41-RR016292 for the ACERT Center Grant to J.F. This work is based upon research conducted at the Advanced Photon Source (APS), Argonne National Laboratory, on the Northeastern Collaborative Access Team beamlines, which are supported by award RR-15301 from the US National Center for Research Resources at the US National Institutes of Health. Use of the APS is supported by the US Department of Energy, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357.
Author information
Authors and Affiliations
Contributions
Y.Z. determined the crystal structure of iron-free PhDph2, X.Z. performed the biochemical studies and prepared protein samples for spectroscopic and structural studies, A.T.T. determined the crystal structure of anaerobically purified PhDph2, M.L. and C.K. performed the Mössbauer spectroscopy, B.D. and J.F. performed the EPR spectroscopy, R.M.K. prepared the initial PhDph2 crystals, E.W. prepared the PhEF2 mutant proteins, S.E.E. supervised the crystallographic studies, H.L. supervised the biochemical studies and H.L., S.E.E. and C.K. prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Table 1 and Supplementary Figures 1-9 with legends. (PDF 9956 kb)
Rights and permissions
About this article
Cite this article
Zhang, Y., Zhu, X., Torelli, A. et al. Diphthamide biosynthesis requires an organic radical generated by an iron–sulphur enzyme. Nature 465, 891–896 (2010). https://doi.org/10.1038/nature09138
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature09138
This article is cited by
-
Monitoring Fe–S cluster occupancy across the E. coli proteome using chemoproteomics
Nature Chemical Biology (2023)
-
LimF is a versatile prenyltransferase for histidine-C-geranylation on diverse non-natural substrates
Nature Catalysis (2022)
-
Iron–sulfur clusters as inhibitors and catalysts of viral replication
Nature Chemistry (2022)
-
Translational fidelity and growth of Arabidopsis require stress-sensitive diphthamide biosynthesis
Nature Communications (2022)
-
QM/MM Study of the Mechanism of the Noncanonical S-Cγ Bond Scission in S-Adenosylmethionine Catalyzed by the CmnDph2 Radical Enzyme
Topics in Catalysis (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.