Abstract
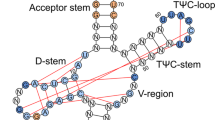
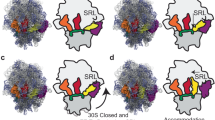
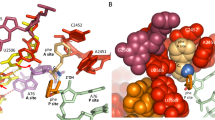
In termination of protein synthesis, the bacterial release factors RF1 and RF2 bind to the ribosome through specific recognition of messenger RNA stop codons and trigger hydrolysis of the bond between the nascent polypeptide and the transfer RNA at the peptidyl-tRNA site, thereby releasing the newly synthesized protein. The release factors are highly specific for a U in the first stop-codon position1 and recognize different combinations of purines in the second and third positions, with RF1 reading UAA and UAG and RF2 reading UAA and UGA. With recently determined crystal structures of termination complexes2,3,4, it has become possible to decipher the energetics of stop-codon reading by computational analysis and to clarify the origin of the high release-factor binding accuracy. Here we report molecular dynamics free-energy calculations on different cognate and non-cognate termination complexes. The simulations quantitatively explain the basic principles of decoding in all three codon positions and reveal the key elements responsible for specificity of the release factors. The overall reading mechanism involves hitherto unidentified interactions and recognition switches that cannot be described in terms of a tripeptide anticodon model. Further simulations of complexes with tRNATrp, the tRNA recognizing the triplet codon for Trp, explain the observation of a ‘leaky’ stop codon5 and highlight the fundamentally different third position reading by RF2, which leads to a high stop-codon specificity with strong discrimination against the Trp codon. The simulations clearly illustrate the versatility of codon reading by protein, which goes far beyond tRNA mimicry.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Freistroffer, D. V., Kwiatkowski, M., Buckingham, R. H. & Ehrenberg, M. The accuracy of codon recognition by polypeptide release factors. Proc. Natl Acad. Sci. USA 97, 2046–2051 (2000)
Laurberg, M. et al. Structural basis for translation termination on the 70S ribosome. Nature 454, 852–857 (2008)
Weixlbaumer, A. et al. Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322, 953–956 (2008)
Korostelev, A. et al. Crystal structure of a translation termination complex formed with release factor RF2. Proc. Natl Acad. Sci. USA 105, 19684–19689 (2008)
Parker, J. Errors and alternatives in reading the universal genetic-code. Microbiol. Rev. 53, 273–298 (1989)
Jørgensen, F., Adamski, F. M., Tate, W. P. & Kurland, C. G. Release factor-dependent false stops are infrequent in Escherichia coli . J. Mol. Biol. 230, 41–50 (1993)
Thompson, R. C. & Stone, P. J. Proofreading of codon-anticodon interaction on ribosomes. Proc. Natl Acad. Sci. USA 74, 198–202 (1977)
Ruusala, T., Ehrenberg, M. & Kurland, C. G. Is there proofreading during polypeptide-synthesis. EMBO J. 1, 741–745 (1982)
Nissen, P. et al. Crystal structure of the ternary complex of Phe-tRNA, EF-Tu, and a GTP analog. Science 270, 1464–1472 (1995)
Kristensen, O., Laurberg, M., Liljas, A. & Selmer, M. Is tRNA binding or tRNA mimicry mandatory for translation factors? Curr. Protein Pept. Sci. 3, 133–141 (2002)
Ito, K., Uno, M. & Nakamura, Y. A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature 403, 680–684 (2000)
Frolova, L. Y. et al. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5, 1014–1020 (1999)
Rawat, U. B. S. et al. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature 421, 87–90 (2003)
Klaholz, B. P. et al. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature 421, 90–94 (2003)
Petry, S. et al. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 123, 1255–1266 (2005)
Trobro, S. & Åqvist, J. A model for how ribosomal release factors induce peptidyl-tRNA cleavage in termination of protein synthesis. Mol. Cell 27, 758–766 (2007)
Ogle, J. M. et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001)
Almlöf, M., Andér, M. & Åqvist, J. Energetics of codon-anticodon recognition on the small ribosomal subunit. Biochemistry 46, 200–209 (2007)
Uno, M., Ito, K. & Nakamura, Y. Polypeptide release at sense and noncognate stop codons by localized charge-exchange alterations in translational release factors. Proc. Natl Acad. Sci. USA 99, 1819–1824 (2002)
Cochella, L. & Green, R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science 308, 1178–1180 (2005)
Hirsh, D. & Gold, L. Translation of UGA triplet in vitro by tryptophan transfer RNAs. J. Mol. Biol. 58, 459–468 (1971)
Zavialov, A. V., Mora, L., Buckingham, R. H. & Ehrenberg, M. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol. Cell 10, 789–798 (2002)
Dong, H., Nilsson, L. & Kurland, C. G. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260, 649–663 (1996)
Adamski, F. M., Mccaughan, K. K., Jorgensen, F., Kurland, C. G. & Tate, W. P. The concentration of polypeptide-chain release factor-1 and factor-2 at different growth-rates of Escherichia coli . J. Mol. Biol. 238, 302–308 (1994)
Bennett, C. H. Efficient estimation of free energy differences from Monte Carlo data. J. Comput. Phys. 22, 245–268 (1976)
Marelius, J., Kolmodin, K., Feierberg, I. & Åqvist, J. Q: a molecular dynamics program for free energy calculations and empirical valence bond simulations in biomolecular systems. J. Mol. Graph. Model. 16, 213–225, 261 (1998)
MacKerell, A. D. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998)
Trobro, S. & Åqvist, J. Mechanism of the translation termination reaction on the ribosome. Biochemistry 48, 11296–11303 (2009)
MacKerell, A. D., Wiorkiewicz-Kuczera, J. & Karplus, M. An all-atom empirical energy function for the simulation of nucleic-acids. J. Am. Chem. Soc. 117, 11946–11975 (1995)
King, G. & Warshel, A. A surface constrained all-atom solvent model for effective simulations of polar solutions. J. Chem. Phys. 91, 3647–3661 (1989)
Trobro, S. & Åqvist, J. Mechanism of peptide bond synthesis on the ribosome. Proc. Natl Acad. Sci. USA 102, 12395–12400 (2005)
Lee, F. S. & Warshel, A. A local reaction field method for fast evaluation of long-range electrostatic interactions in molecular simulations. J. Chem. Phys. 97, 3100–3107 (1992)
Ryckaert, J.-P., Ciccotti, G. & Berendsen, H. J. C. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 (1977)
Acknowledgements
Support from the Swedish Research Council is gratefully acknowledged. We thank M. Ehrenberg for discussions and M. Laurberg, H. Noller, A. Weixlbaumer and V. Ramakrishnan for sending us their atomic coordinates before release and for providing electron density maps.
Author information
Authors and Affiliations
Contributions
J.S. and M.A. performed the simulations. All authors analysed the data and prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Table 1 and Supplementary Figures 1-4 with legends. (PDF 551 kb)
Rights and permissions
About this article
Cite this article
Sund, J., Andér, M. & Åqvist, J. Principles of stop-codon reading on the ribosome. Nature 465, 947–950 (2010). https://doi.org/10.1038/nature09082
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09082
This article is cited by
-
Origin of the omnipotence of eukaryotic release factor 1
Nature Communications (2017)
-
Codon-reading specificities of mitochondrial release factors and translation termination at non-standard stop codons
Nature Communications (2013)
-
Energetics of activation of GTP hydrolysis on the ribosome
Nature Communications (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.