Abstract
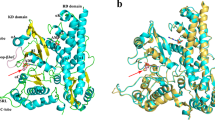
The Escherichia coli isocitrate dehydrogenase kinase/phosphatase (AceK) is a unique bifunctional enzyme that phosphorylates or dephosphorylates isocitrate dehydrogenase (ICDH) in response to environmental changes, resulting in the inactivation or, respectively, activation of ICDH1. ICDH inactivation short-circuits the Krebs cycle by enabling the glyoxlate bypass2,3. It was the discovery of AceK and ICDH that established the existence of protein phosphorylation regulation in prokaryotes1,4. As a 65-kDa protein, AceK is significantly larger than typical eukaryotic protein kinases. Apart from the ATP-binding motif, AceK does not share sequence homology with any eukaryotic protein kinase or phosphatase5,6. Most intriguingly, AceK possesses the two opposing activities of protein kinase and phosphatase within one protein, and specifically recognizes only intact ICDH7,8. Additionally, AceK has strong ATPase activity9. It has been shown that AceK kinase, phosphatase and ATPase activities reside at the same site6,10, although the molecular basis of such multifunctionality and its regulation remains completely unknown. Here we report the structures of AceK and its complex with ICDH. The AceK structure reveals a eukaryotic protein-kinase-like domain containing ATP and a regulatory domain with a novel fold. As an AceK phosphatase activator and kinase inhibitor, AMP is found to bind in an allosteric site between the two AceK domains. An AMP-mediated conformational change exposes and shields ATP, acting as a switch between AceK kinase and phosphatase activities, and ICDH-binding induces further conformational change for AceK activation. The substrate recognition loop of AceK binds to the ICDH dimer, allowing higher-order substrate recognition and interaction, and inducing critical conformational change at the phosphorylation site of ICDH.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
LaPorte, D. C. & Koshland, D. E. Jr. A protein with kinase and phosphatase activities involved in regulation of tricarboxylic acid cycle. Nature 300, 458–460 (1982)
Garnak, M. & Reeves, H. C. Purification and properties of phosphorylated isocitrate dehydrogenase of Escherichia coli . J. Biol. Chem. 254, 7915–7920 (1979)
Hoyt, J. C. & Reeves, H. C. In vivo phosphorylation of isocitrate lyase from Escherichia coli D5H3G7. Biochem. Biophys. Res. Commun. 153, 875–880 (1988)
Garnak, M. & Reeves, H. C. Phosphorylation of isocitrate dehydrogenase of Escherichia coli . Science 203, 1111–1112 (1979)
Klumpp, D. J. et al. Nucleotide sequence of aceK, the gene encoding isocitrate dehydrogenase kinase/phosphatase. J. Bacteriol. 170, 2763–2769 (1988)
Stueland, C. S., Ikeda, T. P. & LaPorte, D. C. Mutation of the predicted ATP binding site inactivates both activities of isocitrate dehydrogenase kinase/phosphatase. J. Biol. Chem. 264, 13775–13779 (1989)
LaPorte, D. C. & Koshland, D. E. Jr. Phosphorylation of isocitrate dehydrogenase as a demonstration of enhanced sensitivity in covalent regulation. Nature 305, 286–290 (1983)
McKee, J. S., Hlodan, R. & Nimmo, H. G. Studies of the phosphorylation of Escherichia coli isocitrate dehydrogenase: recognition of the enzyme by isocitrate dehydrogenase kinase/phosphatase and effects of phosphorylation on its structure and properties. Biochimie 71, 1059–1064 (1989)
Stueland, C. S., Eck, K. R., Stieglbauer, K. T. & LaPorte, D. C. Isocitrate dehydrogenase kinase/phosphatase exhibits an intrinsic adenosine triphosphatase activity. J. Biol. Chem. 262, 16095–16099 (1987)
Miller, S. P., Karschnia, E. J., Ikeda, T. P. & LaPorte, D. C. Isocitrate dehydrogenase kinase/phosphatase: kinetic characteristics of the wild-type and two mutant proteins. J. Biol. Chem. 271, 19124–19128 (1996)
Kornberg, H. L. The role and control of the glyoxylate cycle in Escherichia coli . Biochem. J. 99, 1–11 (1966)
LaPorte, D. C. The isocitrate dehydrogenase phosphorylation cycle: regulation and enzymology. J. Cell. Biochem. 51, 14–18 (1993)
Miller, S. P. et al. Locations of the regulatory sites for isocitrate dehydrogenase kinase/phosphatase. J. Biol. Chem. 275, 833–839 (2000)
LaPorte, D. C., Stueland, C. S. & Ikeda, T. P. Isocitrate dehydrogenase kinase/phosphatase. Biochimie 71, 1051–1057 (1989)
Takio, K. et al. Guanosine cyclic 3′,5′-phosphate dependent protein kinase, a chimeric protein homologous with two separate protein families. Biochemistry 23, 4207–4218 (1984)
Townley, R. & Shapiro, L. Crystal structures of the adenylate sensor from fission yeast AMP-activated protein kinase. Science 315, 1726–1729 (2007)
Kim, C., Xuong, N. H. & Taylor, S. S. Crystal structure of a complex between the catalytic and regulatory (RIα) subunits of PKA. Science 307, 690–696 (2005)
Chen, H. et al. A crystallographic snapshot of tyrosine trans-phosphorylation in action. Proc. Natl Acad. Sci. USA 105, 19660–19665 (2008)
Hurley, J. H. et al. Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase. Proc. Natl Acad. Sci. USA 86, 8635–8639 (1989)
Hurley, J. H., Dean, A. M., Sohl, J. L., Koshland, D. E. Jr & Stroud, R. M. Regulation of an enzyme by phosphorylation at the active site. Science 249, 1012–1016 (1990)
Finer-Moore, J. et al. Access to phosphorylation in isocitrate dehydrogenase may occur by domain shifting. Biochemistry 36, 13890–13896 (1997)
Bennett, P. M. & Holms, W. H. Reversible inactivation of the isocitrate dehydrogenase of Escherichia coli ML308 during growth on acetate. J. Gen. Microbiol. 87, 37–51 (1975)
Ubersax, J. A. & Ferrell, J. E. Jr. Mechanisms of specificity in protein phosphorylation. Nature Rev. Mol. Cell Biol. 8, 530–541 (2007)
Dar, A. C., Dever, T. E. & Sicheri, F. Higher-order substrate recognition of eIF2α by the RNA-dependent protein kinase PKR. Cell 122, 887–900 (2005)
Singh, S. K., Matsuno, K., LaPorte, D. C. & Banaszak, L. J. Crystal structure of Bacillus subtilis isocitrate dehydrogenase at 1.55 Å: insights into the nature of substrate specificity exhibited by Escherichia coli isocitrate dehydrogenase kinase/phosphatase. J. Biol. Chem. 276, 26154–26163 (2001)
Singh, S. K., Miller, S. P., Dean, A., Banaszak, L. J. & LaPorte, D. C. Bacillus subtilis isocitrate dehydrogenase: a substrate analogue for Escherichia coli isocitrate dehydrogenase kinase/phosphatase. J. Biol. Chem. 277, 7567–7573 (2002)
Ikeda, T. & LaPorte, D. C. Isocitrate dehydrogenase kinase/phosphatase: aceK alleles that express kinase but not phosphatase activity. J. Bacteriol. 173, 1801–1806 (1991)
Zheng, J., Lee, D. C. & Jia, Z. Purification, crystallization and preliminary X-ray analysis of isocitrate dehydrogenase kinase/phosphatase from Escherichia coli . Acta Crystallogr. F 65, 536–539 (2009)
Zheng, J., Ji, A. X. & Jia, Z. Purification, crystallization and preliminary X-ray analysis of bifunctional isocitrate dehydrogenase kinase/phosphatase in complex with its substrate, isocitrate dehydrogenase, from Escherichia coli . Acta Crystallogr. F 65, 1153–1156 (2009)
Stueland, C. S., Gorden, K. & LaPorte, D. C. The isocitrate dehydrogenase phosphorylation cycle: identification of the primary rate-limiting step. J. Biol. Chem. 263, 19475–19479 (1988)
Terwilliger, T. C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999)
Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 (2007)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
McRee, D. E. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125, 156–165 (1999)
Brünger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Vagin, A. A. et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D 60, 2184–2195 (2004)
Collaborative. Computation Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Storoni, L. C., McCoy, A. J. & Read, R. J. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D 60, 432–438 (2004)
Holm, L. & Sander, C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995)
Acknowledgements
The authors would like to thank M. Cygler and A. Matte for help in cloning, L. Li and R. Theiss for help in mutagenesis and activity measurements, and J. Allingham and G. Cote for critical reading of the manuscript. We would also like to thank the staff of the Cornell High Energy Synchrotron Source for helping with the collection of synchrotron X-ray data. This work was supported by the Canadian Institutes of Health Research (Z.J.). Z.J. is a Canada Research Chair in Structural Biology.
Author information
Authors and Affiliations
Contributions
Experiments were performed by J.Z. Data were analysed by J.Z. and Z.J. The manuscript was prepared by J.Z. and Z.J.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Results and Discussion, Supplementary Tables 1-5, Supplementary Figures 1-6 with legends and References. (PDF 2205 kb)
Supplementary Movie 1
This movie shows the interaction of AceK with ICDH. ICHD dimer is coloured in marine blue. AceK is coloured in green. The phosphorylation site is coloured in gold. (MOV 8108 kb)
Rights and permissions
About this article
Cite this article
Zheng, J., Jia, Z. Structure of the bifunctional isocitrate dehydrogenase kinase/phosphatase. Nature 465, 961–965 (2010). https://doi.org/10.1038/nature09088
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09088
This article is cited by
-
Transcriptomics analyses and biochemical characterization of Aspergillus flavus spores exposed to 1-nonanol
Applied Microbiology and Biotechnology (2022)
-
Structure and allosteric regulation of human NAD-dependent isocitrate dehydrogenase
Cell Discovery (2020)
-
Characterization of metal binding of bifunctional kinase/phosphatase AceK and implication in activity modulation
Scientific Reports (2019)
-
Evaluation of the Potential Phosphorylation Effect on Isocitrate Dehydrogenases from Saccharomyces cerevisiae and Yarrowia lipolytica
Applied Biochemistry and Biotechnology (2019)
-
Studies on the activation of isocitrate dehydrogenase kinase/phosphatase (AceK) by Mn2+ and Mg2+
BioMetals (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.