Abstract
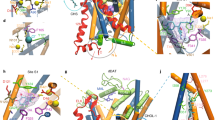
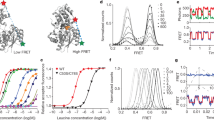
Neurotransmitter:Na+ symporters (NSS) remove neurotransmitters from the synapse in a reuptake process that is driven by the Na+ gradient. Drugs that interfere with this reuptake mechanism, such as cocaine and antidepressants, profoundly influence behaviour and mood. To probe the nature of the conformational changes that are associated with substrate binding and transport, we have developed a single-molecule fluorescence imaging assay and combined it with functional and computational studies of the prokaryotic NSS homologue LeuT. Here we show molecular details of the modulation of intracellular gating of LeuT by substrates and inhibitors, as well as by mutations that alter binding, transport or both. Our direct observations of single-molecule transitions, reflecting structural dynamics of the intracellular region of the transporter that might be masked by ensemble averaging or suppressed under crystallographic conditions, are interpreted in the context of an allosteric mechanism that couples ion and substrate binding to transport.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Amara, S. G. & Sonders, M. S. Neurotransmitter transporters as molecular targets for addictive drugs. Drug Alcohol Depend. 51, 87–96 (1998)
Rudnick, G. Mechanisms of Biogenic Amine Neurotransmitter Transporters 2nd edn (Humana, Totowa, New Jersey, 2002)
Sonders, M. S., Quick, M. & Javitch, J. A. How did the neurotransmitter cross the bilayer? A closer view. Curr. Opin. Neurobiol. 15, 296–304 (2005)
Gu, H., Wall, S. C. & Rudnick, G. Stable expression of biogenic amine transporters reveals differences in inhibitor sensitivity, kinetics, and ion dependence. J. Biol. Chem. 269, 7124–7130 (1994)
Torres, G. E., Gainetdinov, R. R. & Caron, M. G. Plasma membrane monoamine transporters: structure, regulation and function. Nature Rev. Neurosci. 4, 13–25 (2003)
Krause, S. & Schwarz, W. Identification and selective inhibition of the channel mode of the neuronal GABA transporter 1. Mol. Pharmacol. 68, 1728–1735 (2005)
Iversen, L. Neurotransmitter transporters and their impact on the development of psychopharmacology. Br. J. Pharmacol. 147 (suppl. 1). S82–S88 (2006)
Beuming, T., Shi, L., Javitch, J. A. & Weinstein, H. A comprehensive structure-based alignment of prokaryotic and eukaryotic neurotransmitter/Na+ symporters (NSS) aids in the use of the LeuT structure to probe NSS structure and function. Mol. Pharmacol. 70, 1630–1642 (2006)
Yamashita, A. et al. Crystal structure of a bacterial homologue of Na+/Cl-dependent neurotransmitter transporters. Nature 437, 215–223 (2005)
Shi, L. et al. The mechanism of a neurotransmitter:sodium symporter—inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol. Cell 30, 667–677 (2008)
Zhou, Z. et al. LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science 317, 1390–1393 (2007)
Singh, S. K., Yamashita, A. & Gouaux, E. Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature 448, 952–956 (2007)
Zhou, Z. et al. Antidepressant specificity of serotonin transporter suggested by three LeuT-SSRI structures. Nature Struct. Mol. Biol. 16, 652–657 (2009)
Singh, S. K., Piscitelli, C. L., Yamashita, A. & Gouaux, E. A competitive inhibitor traps LeuT in an open-to-out conformation. Science 322, 1655–1661 (2008)
Quick, M. et al. Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor-bound conformation. Proc. Natl Acad. Sci. USA 106, 5563–5568 (2009)
Blanchard, S. C. Single-molecule observations of ribosome function. Curr. Opin. Struct. Biol. 19, 103–109 (2009)
Kinosita, K. Jr, Yasuda, R. & Noji, H. F1-ATPase: a highly efficient rotary ATP machine. Essays Biochem. 35, 3–18 (2000)
Vale, R. D. Myosin V motor proteins: marching stepwise towards a mechanism. J. Cell Biol. 163, 445–450 (2003)
Peterman, E. J., Sosa, H. & Moerner, W. E. Single-molecule fluorescence spectroscopy and microscopy of biomolecular motors. Annu. Rev. Phys. Chem. 55, 79–96 (2004)
Ishii, Y., Nishiyama, M. & Yanagida, T. Mechano-chemical coupling of molecular motors revealed by single molecule measurements. Curr. Protein Pept. Sci. 5, 81–87 (2004)
Zhuang, X. Single-molecule RNA science. Annu. Rev. Biophys. Biomol. Struct. 34, 399–414 (2005)
Cornish, P. V. & Ha, T. A survey of single-molecule techniques in chemical biology. ACS Chem. Biol. 2, 53–61 (2007)
Park, H., Toprak, E. & Selvin, P. R. Single-molecule fluorescence to study molecular motors. Q. Rev. Biophys. 40, 87–111 (2007)
Ha, T. Need for speed: mechanical regulation of a replicative helicase. Cell 129, 1249–1250 (2007)
Schuler, B. & Eaton, W. A. Protein folding studied by single-molecule FRET. Curr. Opin. Struct. Biol. 18, 16–26 (2008)
Herbert, K. M., Greenleaf, W. J. & Block, S. M. Single-molecule studies of RNA polymerase: motoring along. Annu. Rev. Biochem. 77, 149–176 (2008)
Wen, J. D. et al. Following translation by single ribosomes one codon at a time. Nature 452, 598–603 (2008)
Pyle, A. M. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys 37, 317–336 (2008)
Noskov, S. Y. & Roux, B. Control of ion selectivity in LeuT: two Na+ binding sites with two different mechanisms. J. Mol. Biol. 377, 804–818 (2008)
Celik, L., Schiott, B. & Tajkhorshid, E. Substrate binding and formation of an occluded state in the leucine transporter. Biophys. J. 94, 1600–1612 (2008)
Jorgensen, A. M., Tagmose, L., Bogeso, K. P. & Peters, G. H. Molecular dynamics simulations of Na+/Cl--dependent neurotransmitter transporters in a membrane-aqueous system. ChemMedChem 2, 827–840 (2007)
Kniazeff, J. et al. An intracellular interaction network regulates conformational transitions in the dopamine transporter. J. Biol. Chem. 283, 17691–17701 (2008)
Caplan, D. A., Subbotina, J. O. & Noskov, S. Y. Molecular mechanism of ion-ion and ion-substrate coupling in the Na+-dependent leucine transporter LeuT. Biophys. J. 95, 4613–4621 (2008)
Munro, J. B., Altman, R. B., O'Connor, N. & Blanchard, S. C. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell 25, 505–517 (2007)
Roy, R., Hohng, S. & Ha, T. A practical guide to single-molecule FRET. Nature Methods 5, 507–516 (2008)
Dave, R., Terry, D. S., Munro, J. B. & Blanchard, S. C. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys. J. 96, 2371–2381 (2009)
Bennett, E. R., Su, H. & Kanner, B. I. Mutation of arginine 44 of GAT-1, a (Na+ + Cl-)-coupled gamma-aminobutyric acid transporter from rat brain, impairs net flux but not exchange. J. Biol. Chem. 275, 34106–34113 (2000)
Shaffer, P. L., Goehring, A., Shankaranarayanan, A. & Gouaux, E. Structure and mechanism of a Na+-independent amino acid transporter. Science 325, 1010–1014 (2009)
Lockless, S. W. & Ranganathan, R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science 286, 295–299 (1999)
Beckett, D., Kovaleva, E. & Schatz, P. J. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8, 921–929 (1999)
Majumdar, D. S. et al. Single-molecule FRET reveals sugar-induced conformational dynamics in LacY. Proc. Natl Acad. Sci. USA 104, 12640–12645 (2007)
Nie, Y., Sabetfard, F. E. & Kaback, H. R. The Cys154→Gly mutation in LacY causes constitutive opening of the hydrophilic periplasmic pathway. J. Mol. Biol. 379, 695–703 (2008)
Quick, M. & Javitch, J. A. Monitoring the function of membrane transport proteins in detergent-solubilized form. Proc. Natl Acad. Sci. USA 104, 3603–3608 (2007)
Schaffner, W. & Weissmann, C. A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal. Biochem. 56, 502–514 (1973)
Lakowicz, J. R. Principles of Fluorescence Spectroscopy 3rd edn (Springer, 2006)
Mujumdar, R. B. et al. Cyanine dye labeling reagents: sulfoindocyanine succinimidyl esters. Bioconjug. Chem. 4, 105–111 (1993)
Williams, A., Winfield, A. S. & Miller, N. J. Relative fluorescence quantum yields using a computer-controlled luminescence spectrometer. Analyst (Lond.) 108, 1067–1071 (1983)
Karstens, T. & Kobs, K. Rhodamine B and rhodamine 101 as reference substances for fluorescence quantum yield measurements. J. Phys. Chem. 84, 1871–1872 (1980)
Blanchard, S. C. et al. tRNA dynamics on the ribosome during translation. Proc. Natl Acad. Sci. USA 101, 12893–12898 (2004)
Qin, F. Restoration of single-channel currents using the segmental k-means method based on hidden Markov modeling. Biophys. J. 86, 1488–1501 (2004)
Qin, F., Auerbach, A. & Sachs, F. Estimating single-channel kinetic parameters from idealized patch-clamp data containing missed events. Biophys. J. 70, 264–280 (1996)
Acknowledgements
We thank R. Altman for assistance with reagents for single-molecule experiments; F. Carvalho for the preparation of membranes; J. Munro for help measuring anisotropy; R. Dave for preliminary photobleaching optimization studies; and M. Quick for helpful discussion and comments on the manuscript. Molecular graphic figures and movies were prepared with PyMOL (DeLano Scientific; http://www.pymol.org). Computations were performed on Ranger at the Texas Advanced Computing Center (TG-MCB090022) and the David A. Cofrin computational infrastructure of the Institute for Computational Biomedicine at Weill Cornell Medical College. This work was supported in part by National Institutes of Health Grants DA17293 and DA022413 (J.A.J.), DA12408 (H.W.), and DA023694 (L.S.). D.S.T. is supported by the Tri-Institutional Training Program in Computational Biology and Medicine.
Author information
Authors and Affiliations
Contributions
Y.Z. expressed, purified, labelled and functionally characterized the LeuT mutants. Y.Z. and D.T. designed, carried out, and analysed the single-molecule experiments. L.S. and H.W. designed and analysed the computational studies, which were carried out by L.S., S.C.B. and J.A.J. helped to design the biochemical and single-molecule experiments and, with L.S. and H.W., helped to interpret the data. All the authors contributed to writing and editing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion, Supplementary Figures 1-8 with legends, Supplementary Tables 1-3, Notes for Supplementary Movies 1-2 and a Reference. (PDF 1324 kb)
Supplementary Movie 1
This flash movie shows a side view of conformational changes in the simulated transport mechanism from a ~180 ns molecular dynamics simulation that induces and equilibrates the inward-open structure (orange) starting from the LeuT crystal structure (gray). The conformational changes agree with and are reflected in the results from the smFRET measurements. (SWF 9327 kb)
Supplementary Movie 2
This flash movie shows an intracellular view of conformational changes in the simulated transport mechanism from a ~180 ns molecular dynamics simulation that induces and equilibrates the inward-open structure (orange) starting from the LeuT crystal structure (gray). The conformational changes agree with and are reflected in the results from the smFRET measurements. (SWF 9399 kb)
Rights and permissions
About this article
Cite this article
Zhao, Y., Terry, D., Shi, L. et al. Single-molecule dynamics of gating in a neurotransmitter transporter homologue. Nature 465, 188–193 (2010). https://doi.org/10.1038/nature09057
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature09057
This article is cited by
-
Autoregulation of GPCR signalling through the third intracellular loop
Nature (2023)
-
Multi-parameter photon-by-photon hidden Markov modeling
Nature Communications (2022)
-
On the physical mechanisms underlying single molecule dynamics in simple liquids
Scientific Reports (2021)
-
Single-molecule FRET imaging of GPCR dimers in living cells
Nature Methods (2021)
-
X-ray structure of LeuT in an inward-facing occluded conformation reveals mechanism of substrate release
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.