Abstract
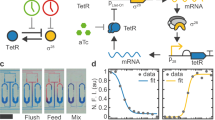
The engineering of genetic circuits with predictive functionality in living cells represents a defining focus of the expanding field of synthetic biology. This focus was elegantly set in motion a decade ago with the design and construction of a genetic toggle switch and an oscillator, with subsequent highlights that have included circuits capable of pattern generation, noise shaping, edge detection and event counting. Here we describe an engineered gene network with global intercellular coupling that is capable of generating synchronized oscillations in a growing population of cells. Using microfluidic devices tailored for cellular populations at differing length scales, we investigate the collective synchronization properties along with spatiotemporal waves occurring at millimetre scales. We use computational modelling to describe quantitatively the observed dependence of the period and amplitude of the bulk oscillations on the flow rate. The synchronized genetic clock sets the stage for the use of microbes in the creation of a macroscopic biosensor with an oscillatory output. Furthermore, it provides a specific model system for the generation of a mechanistic description of emergent coordinated behaviour at the colony level.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Huygens, C. Œuvres complètes de Christiaan Huygens Vol. 17 (Martinus Nijhoff, The Hague, 1932)
Pikovsky, A., Rosenblum, M. & Kurths, J. Synchronization: A Universal Concept in Nonlinear Sciences (Cambridge, 2001)
Strogatz, S. Sync (Penguin Books New York, 2004)
Vladimirov, A. G., Kozyreff, G. & Mandel, P. Synchronization of weakly stable oscillators and semiconductor laser arrays. Europhys. Lett. 61, 613–619 (2003)
Wiesenfeld, K., Colet, P. & Strogatz, S. Synchronization transitions in a disordered Josephson series array. Phys. Rev. Lett. 76, 404–407 (1996)
Lewandowski, W., Azoubib, J. & Klepczynski, W. GPS: primary tool for time transfer. Proc. IEEE 87, 163–172 (1999)
Li, D., Wong, K., Hu, Y. & Sayeed, A. Detection, classification and tracking of targets in distributed sensor networks. IEEE Signal Process. Mag. 19, 17–29 (2002)
Winfree, A. T. Biological rhythms and the behavior of populations of coupled oscillators. J. Theor. Biol. 16, 15–42 (1967)
Mirollo, R. & Strogatz, S. Synchronization of pulse-coupled biological oscillators. SIAM J. Appl. Math. 50, 1645–1662 (1990)
Elson, R. C. et al. Synchronous behavior of two coupled biological neurons. Phys. Rev. Lett. 81, 5692–5695 (1998)
Jiang, Y.-J. et al. Notch signalling and the synchronization of the somite segmentation clock. Nature 408, 475–479 (2000)
Glass, L. Synchronization and rhythmic processes in physiology. Nature 410, 277–284 (2001)
Young, M. W. & Kay, S. Time zones: a comparative genetics of circadian clocks. Nature Rev. Genet. 2, 702–715 (2001)
Chabot, J. R., Pedraza, J., Luitel, P. & van Oudenaarden, A. Stochastic gene expression out-of-steady-state in the cyanobacterial circadian clock. Nature 450, 1249–1252 (2007)
Kerckhoffs, R. C. P., McCulloch, A., Omens, J. & Mulligan, L. Effects of biventricular pacing and scar size in a computational model of the failing heart with left bundle branch block. Med. Image Anal. 13, 362–369 (2009)
Grenier, F., Timofeev, I. & Steriade, M. Neocortical very fast oscillations (ripples, 80–200 Hz) during seizures: intracellular correlates. J. Neurophysiol. 89, 841–852 (2003)
Isalan, M. et al. Evolvability and hierarchy in rewired bacterial gene networks. Nature 452, 840–845 (2008)
Alon, U. Network motifs: theory and experimental approaches. Nature Rev. Genet. 8, 450–461 (2007)
Gibson, D. G. et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319, 1215–1220 (2008)
Hasty, J., McMillen, D. & Collins, J. J. Engineered gene circuits. Nature 420, 224–230 (2002)
Endy, D. Foundations for engineering biology. Nature 438, 449–453 (2005)
Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli . Nature 403, 339–342 (2000)
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000)
You, L., Cox, R., Weiss, R. & Arnold, F. Programmed population control by cell–cell communication and regulated killing. Nature 428, 868–871 (2004)
Basu, S., Gerchman, Y., Collins, C., Arnold, F. & Weiss, R. A synthetic multicellular system for programmed pattern formation. Nature 434, 1130–1134 (2005)
Kobayashi, H. et al. Programmable cells: interfacing natural and engineered gene networks. Proc. Natl Acad. Sci. USA 101, 8414–8419 (2004)
Austin, D. W. et al. Gene network shaping of inherent noise spectra. Nature 439, 608–611 (2006)
Tabor, J. J. et al. A synthetic genetic edge detection program. Cell 137, 1272–1281 (2009)
Friedland, A. E. et al. Synthetic gene networks that count. Science 324, 1199–1202 (2009)
Atkinson, M. R., Savageau, M. A., Myers, J. T. & Ninfa, A. J. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli . Cell 113, 597–607 (2003)
Stricker, J. et al. A fast, robust and tunable synthetic gene oscillator. Nature 456, 516–519 (2008)
Tigges, M., Marquez-Lago, T., Stelling, J. & Fussenegger, M. A tunable synthetic mammalian oscillator. Nature 457, 309–312 (2009)
Fung, E. et al. A synthetic gene-metabolic oscillator. Nature 435, 118–122 (2005)
Cookson, N. A., Tsimring, L. S. & Hasty, J. The pedestrian watchmaker: Genetic clocks from engineered oscillations. FEBS Lett. 483, 3931–3937 (2009)
Bennett, M. R. & Hasty, J. Microfluidic devices for measuring gene network dynamics in single cells. Nature Rev. Genet. 10, 628–638 (2009)
Waters, C. M. & Bassler, B. L. Quorum sensing: cell-to-cell communication in bacteria. Ann. Rev. Cell Dev. Biol. 21, 319–346 (2005)
Liu, D. et al. Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures. Biochemistry 47, 7706–7714 (2008)
Glossop, N. R. J., Lyons, L. C. & Hardin, P. E. Interlocked feedback loops within the Drosophila circadian oscillator. Science 286, 766–768 (1999)
Lakin-Thomas, P. L. & Brody, S. Circadian rhythms in microorganisms: new complexities. Annu. Rev. Microbiol. 58, 489–519 (2004)
McMillen, D., Kopell, N., Hasty, J. & Collins, J. Synchronizing genetic relaxation oscillators by intercell signaling. Proc. Natl Acad. Sci. USA 99, 679–684 (2002)
Garcia-Ojalvo, J., Elowitz, M. & Strogatz, S. Modeling a synthetic multicellular clock: repressilators coupled by quorum sensing. Proc. Natl Acad. Sci. USA 101, 10955–10960 (2004)
Reading, N. C. & Sperandio, V. Quorum sensing: the many languages of bacteria. FEMS Microbiol. Lett. 254, 1–11 (2006)
Cookson, S., Ostroff, N., Pang, W., Volfson, D. & Hasty, J. Monitoring dynamics of single-cell gene expression over multiple cell cycles. Mol. Syst. Biol. 1, 2005.0024 (2005)
Mather, W., Bennett, M., Hasty, J. & Tsimring, L. Delay-induced degrade-and-fire oscillations in small genetic circuits. Phys. Rev. Lett. 102, 068105 (2009)
Ozbudak, E. M., Thattai, M., Kurtser, I., Grossman, A. D. & van Oudenaarden, A. Regulation of noise in the expression of a single gene. Nature Genet. 31, 69–73 (2002)
Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002)
Lutz, R. & Bujard, H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1–I2 regulatory elements. Nucleic Acids Res. 25, 1203 (1997)
Dunlap, P. V. & Greenberg, E. P. Control of Vibrio fischeri luminescence gene expression in Escherichia coli by cyclic AMP and cyclic AMP receptor protein. J. Bacteriol. 164, 45–50 (1985)
Thomas, P. W. et al. The quorum-quenching lactonase from Bacillus thuringiensis is a metalloprotein. Biochemistry 44, 7559–7569 (2005)
Acknowledgements
We thank J. Stricker for helpful discussions on plasmid construction, and M. Bennett, K. Wiesenfeld and J. Collins for stimulating discussions during the preparation of the manuscript. This work was supported by the National Institutes of Health and General Medicine (GM69811), the DOE CSGF fellowship (to T.D.), and CONACyT (Mexico, grant 184646, to O.M.-P.).
Author Contributions All authors contributed extensively to the work presented in this paper. T.D. and O.M.-P. are equally contributing first authors, and L.T. and J.H. are equally contributing senior authors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Data, Supplementary Figures 1- 6 with Legends and Supplementary References. (PDF 621 kb)
Supplementary Movie 1
This movie shows time lapse fluorescence microscopy of TDQS1 cells at low flow rate in a 100x100μm trap. (MOV 4428 kb)
Supplementary Movie 2
This movie shows time lapse fluorescence microscopy of TDQS1 cells in a 2000x100x0.95μm open trap showing propagation of AHL at millimeter scale. (MOV 2066 kb)
Supplementary Movie 3
This movie shows time lapse microscopy of TDQS1 cells at high flow rate in a 100x100μm trap. (MOV 9496 kb)
Supplementary Movie 4
This movie shows zoomed time lapse fluorescence microscopy of TDQS1 cells in a 2000x100x0.95μm open trap showing close-up of cells and propagation of AHL. (MOV 16235 kb)
Supplementary Movie 5
This movie shows time lapse fluorescence microscopy of TDQS1 cells in a three dimensional 1000x400x4.0μm trap. (MOV 2448 kb)
Supplementary Movie 6
This movie shows simulation of the wave propagation within a uniform population of cells. (MOV 1268 kb)
Supplementary Movie 7
This movie shows simulation of the wave propagation within a growing dense cluster of cells. (MOV 288 kb)
Rights and permissions
About this article
Cite this article
Danino, T., Mondragón-Palomino, O., Tsimring, L. et al. A synchronized quorum of genetic clocks. Nature 463, 326–330 (2010). https://doi.org/10.1038/nature08753
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature08753
This article is cited by
-
A synthetic population-level oscillator in non-microfluidic environments
Communications Biology (2023)
-
De novo engineering of a bacterial lifestyle program
Nature Chemical Biology (2023)
-
Fitness cost associated with cell phenotypic switching drives population diversification dynamics and controllability
Nature Communications (2023)
-
Programming bacteria for multiplexed DNA detection
Nature Communications (2023)
-
Engineering the gut microbiome
Nature Reviews Bioengineering (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.