Abstract
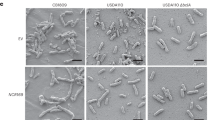
Homocitrate is a component of the iron–molybdenum cofactor in nitrogenase, where nitrogen fixation occurs1,2. NifV, which encodes homocitrate synthase (HCS)3, has been identified from various diazotrophs but is not present in most rhizobial species that perform efficient nitrogen fixation only in symbiotic association with legumes. Here we show that the FEN1 gene of a model legume, Lotus japonicus, overcomes the lack of NifV in rhizobia for symbiotic nitrogen fixation. A Fix- (non-fixing) plant mutant, fen1, forms morphologically normal but ineffective nodules4,5. The causal gene, FEN1, was shown to encode HCS by its ability to complement a HCS-defective mutant of Saccharomyces cerevisiae. Homocitrate was present abundantly in wild-type nodules but was absent from ineffective fen1 nodules. Inoculation with Mesorhizobium loti carrying FEN1 or Azotobacter vinelandii NifV rescued the defect in nitrogen-fixing activity of the fen1 nodules. Exogenous supply of homocitrate also recovered the nitrogen-fixing activity of the fen1 nodules through de novo nitrogenase synthesis in the rhizobial bacteroids. These results indicate that homocitrate derived from the host plant cells is essential for the efficient and continuing synthesis of the nitrogenase system in endosymbionts, and thus provide a molecular basis for the complementary and indispensable partnership between legumes and rhizobia in symbiotic nitrogen fixation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Hoover, T. R. et al. Identification of the V factor needed for synthesis of the iron-molybdenum cofactor of nitrogenase as homocitrate. Nature 329, 855–857 (1987)
Hoover, T. R., Imperial, J., Ludden, P. W. & Shah, V. K. Homocitrate is a component of the iron-molybdenum cofactor of nitrogenase. Biochemistry 28, 2768–2771 (1989)
Zheng, L., White, R. H. & Dean, D. R. Purification of the Azotobacter vinelandii nifV-encoded homocitrate synthase. J. Bacteriol. 179, 5963–5966 (1997)
Imaizumi-Anraku, H. et al. Two ineffective-nodulating mutants of Lotus japonicus—different phenotypes caused by the blockage of endocytotic bacterial release and nodule maturation. Plant Cell Physiol. 38, 871–881 (1997)
Kawaguchi, M. et al. Root, root hair, and symbiotic mutants of the model legume Lotus japonicus . Mol. Plant Microbe Interact. 15, 17–26 (2002)
Asamizu, E., Nakamura, Y., Sato, S. & Tabata, S. Characteristics of the Lotus japonicus gene repertoire deduced from large-scale expressed sequence tag (EST) analysis. Plant Mol. Biol. 54, 405–414 (2004)
Kouchi, H. & Hata, S. GmN56, a novel nodule-specific cDNA from soybean root nodules encodes a protein homologous to isopropylmalate synthase and homocitrate synthase. Mol. Plant Microbe Interact. 8, 172–176 (1995)
Casalone, E., Barberio, C., Cavalieri, D. & Polsinelli, M. Identification by functional analysis of the gene encoding α-isopropylmalate synthase II (LEU9) in Saccharomyces cerevisiae . Yeast 16, 539–545 (2000)
De Kraker, J. W. et al. Two Arabidopsis genes (IPMS1 and IPMS2) encode isopropylmalate synthase, the branchpoint step in the biosynthesis of leucine. Plant Physiol. 143, 970–986 (2007)
Feller, A., Ramos, F., Piérard, A. & Dubois, E. In Saccharomyces cerevisiae, feedback inhibition of homocitrate synthase isoenzymes by lysine modulates the activation of LYS gene expression by Lys14p. Eur. J. Biochem. 261, 163–170 (1999)
Kneip, C., Lockhart, P., Voss, C. & Maier, U. G. Nitrogen fixation in eukaryotes—new models for symbiosis. BMC Evol. Biol. 7, 55–66 (2007)
Dreyfus, B. L., Elmerich, C. & Dommergues, Y. R. Free-living Rhizobium strain able to grow on N2 as the sole nitrogen source. Appl. Environ. Microbiol. 45, 711–713 (1983)
Giraud, E. & Fleischman, D. Nitrogen-fixing symbiosis between photosynthetic bacteria and legumes. Photosynth. Res. 82, 115–130 (2004)
Giraud, E. et al. Legumes symbioses: absence of nod genes in photosynthetic bradyrhizobia. Science 316, 1307–1312 (2007)
Gauthier, D., Diem, H. G. & Dommergues, Y. In vitro nitrogen fixation by two actinomycete strains isolated from Casuarina nodules. Appl. Environ. Microbiol. 41, 306–308 (1981)
Pagan, J. D., Child, J. J., Scowcroft, W. R. & Gibson, A. H. Nitrogen fixation by Rhizobium cultured on a defined medium. Nature 256, 406–407 (1975)
Kurz, W. G. W. & LaRue, T. A. Nitrogenase activity in rhizobia in absence of plant host. Nature 256, 407–409 (1975)
McComb, J. A., Elliott, J. & Dilworth, M. J. Acetylene reduction by Rhizobium in pure culture. Nature 256, 409–410 (1975)
Hoover, T. R. et al. Dinitrogenase with altered substrate specificity results from the use of homocitrate analogues for in vitro synthesis of the iron-molybdenum cofactor. Biochemistry 27, 3647–3652 (1988)
Kohlhaw, G. & Leary, T. R. α-Isopropylmalate synthase from Salmonella typhimurium . J. Biol. Chem. 244, 2218–2225 (1969)
Ulm, E. H., Böhme, R. & Kohlhaw, G. α-Isopropylmalate synthase from yeast: purification, kinetic studies, and effect of ligands on stability. J. Bacteriol. 110, 1118–1126 (1972)
Jacobsent-Lyon, K. et al. Symbiotic and nonsymbiotic hemoglobin genes of Casuarina glauca . Plant Cell 7, 213–223 (1995)
Takane, K., Tajima, S. & Kouchi, H. Two distinct uricase II (nodulin 35) genes are differentially expressed in soybean plants. Mol. Plant Microbe Interact. 6, 735–741 (1997)
Hata, S., Izui, K. & Kouchi, H. Expression of a soybean nodule-enhanced phosphoenolpyruvate carboxylase gene that shows striking similarity to another gene for a house-keeping isoform. Plant J. 13, 267–273 (1998)
Ronson, C. W., Lyttleton, P. & Robertson, J. G. C4-dicarboxylate transport mutants of Rhizobium trifolii form ineffective nodules on Trifolium repens . Proc. Natl Acad. Sci. USA 78, 4284–4288 (1981)
Prell, J. et al. Legumes regulate Rhizobium bacteroid development and persistence by the supply of branched-chain amino acid. Proc. Natl Acad. Sci. USA 106, 12477–12482 (2009)
Sato, S. et al. Genome structure of the legume, Lotus japonicus . DNA Res. 15, 227–239 (2008)
Suganuma, N. et al. The Lotus japonicus Sen1 gene controls rhizobial differentiation into nitrogen-fixing bacteroids in nodules. Mol. Genet. Genomics 269, 312–320 (2003)
Xu, P. F., Matsumoto, T., Ohki, Y. & Tatsumi, K. A facile method for synthesis of (R)-(–)- and (S)-(+)-homocitric acid lactones and related α-hydroxy dicarboxylic acids from D- or L-malic acid. Tetrahedr. Lett. 46, 3815–3818 (2005)
Shimomura, K. et al. LjnsRING, a novel RING finger protein, is required for symbiotic interactions between Mesorhizobium loti and Lotus japonicus . Plant Cell Physiol. 47, 1572–1581 (2006)
Diaz, C. L., Schlaman, H. R. M. & Spaink, H. P. in Lotus japonicus Handbook (Marquez, A. J., ed.) 261–277 (Springer, 2005)
Suganuma, N. et al. cDNA macroarray analysis of gene expression in ineffective nodules induced on the Lotus japonicus sen1 mutant. Mol. Plant Microbe Interact. 17, 1223–1233 (2004)
Wilson, J. K. et al. β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other Gram-negative bacteria. Microbiology 141, 1691–1705 (1995)
Kumagai, H. et al. A novel ankyrin-repeat membrane protein, IGN1, is required for persistence of nitrogen-fixing symbiosis in root nodules of Lotus japonicus . Plant Physiol. 143, 1293–1305 (2007)
Kouchi, H., Fukai, K. & Kihara, A. Metabolism of glutamate and aspartate in bacteroids isolated from soybean root nodules. J. Gen. Microbiol. 137, 2901–2910 (1991)
Acknowledgements
We thank E. Casalone and E. Dubois for providing Saccharomyces cerevisiae mutants; T. Bisseling for providing nitrogenase antibodies; Y. Kawaharada and H. Mitsui for technical help; and R. W. Ridge for critical reading of the manuscript. This work was supported by the Special Coordination Funds for Promoting Science and Technology of the Japanese Ministry of Education, Culture, Sports, Science and Technology.
Author Contributions All authors contributed extensively to the experimental work. The manuscript was written by T.H., H.K. and N.S.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Results, Supplementary Figures 1-6 with Legends and Supplementary Table 1. (PDF 292 kb)
Rights and permissions
About this article
Cite this article
Hakoyama, T., Niimi, K., Watanabe, H. et al. Host plant genome overcomes the lack of a bacterial gene for symbiotic nitrogen fixation. Nature 462, 514–517 (2009). https://doi.org/10.1038/nature08594
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature08594
This article is cited by
-
Symbiosis for rhizobia is not an easy ride
Nature Microbiology (2024)
-
Evolutionary origin and ecological implication of a unique nif island in free-living Bradyrhizobium lineages
The ISME Journal (2021)
-
Control of nitrogen fixation in bacteria that associate with cereals
Nature Microbiology (2019)
-
Rhizobia: from saprophytes to endosymbionts
Nature Reviews Microbiology (2018)
-
Hopanoid lipids: from membranes to plant–bacteria interactions
Nature Reviews Microbiology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.