Abstract
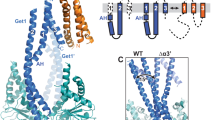
Targeting of newly synthesized membrane proteins to the endoplasmic reticulum is an essential cellular process. Most membrane proteins are recognized and targeted co-translationally by the signal recognition particle. However, nearly 5% of membrane proteins are ‘tail-anchored’ by a single carboxy-terminal transmembrane domain that cannot access the co-translational pathway. Instead, tail-anchored proteins are targeted post-translationally by a conserved ATPase termed Get3. The mechanistic basis for tail-anchored protein recognition or targeting by Get3 is not known. Here we present crystal structures of yeast Get3 in ‘open’ (nucleotide-free) and ‘closed’ (ADP·AlF4--bound) dimer states. In the closed state, the dimer interface of Get3 contains an enormous hydrophobic groove implicated by mutational analyses in tail-anchored protein binding. In the open state, Get3 undergoes a striking rearrangement that disrupts the groove and shields its hydrophobic surfaces. These data provide a molecular mechanism for nucleotide-regulated binding and release of tail-anchored proteins during their membrane targeting by Get3.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
17 September 2009
Author affiliations were added for A.S. and M.D. on 17 September 2009.
References
Egea, P. F., Stroud, R. M. & Walter, P. Targeting proteins to membranes: structure of the signal recognition particle. Curr. Opin. Struct. Biol. 15, 213–220 (2005)
Keenan, R. J., Freymann, D. M., Stroud, R. M. & Walter, P. The signal recognition particle. Annu. Rev. Biochem. 70, 755–775 (2001)
Rapoport, T. A. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450, 663–669 (2007)
Beilharz, T., Egan, B., Silver, P. A., Hofmann, K. & Lithgow, T. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae . J. Biol. Chem. 278, 8219–8223 (2003)
Kalbfleisch, T., Cambon, A. & Wattenberg, B. W. A bioinformatics approach to identifying tail-anchored proteins in the human genome. Traffic 8, 1687–1694 (2007)
Borgese, N., Brambillasca, S. & Colombo, S. How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 19, 368–375 (2007)
Wattenberg, B. & Lithgow, T. Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic 2, 66–71 (2001)
Kutay, U., Ahnert-Hilger, G., Hartmann, E., Wiedenmann, B. & Rapoport, T. A. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 14, 217–223 (1995)
Favaloro, V., Spasic, M., Schwappach, B. & Dobberstein, B. Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J. Cell Sci. 121, 1832–1840 (2008)
Stefanovic, S. & Hegde, R. S. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 128, 1147–1159 (2007)
Kurdi-Haidar, B. et al. Isolation of the ATP-binding human homolog of the arsA component of the bacterial arsenite transporter. Genomics 36, 486–491 (1996)
Kurdi-Haidar, B., Heath, D., Aebi, S. & Howell, S. B. Biochemical characterization of the human arsenite-stimulated ATPase (hASNA-I). J. Biol. Chem. 273, 22173–22176 (1998)
Rosen, B. P., Bhattacharjee, H., Zhou, T. & Walmsley, A. R. Mechanism of the ArsA ATPase. Biochim. Biophys. Acta 1461, 207–215 (1999)
Schuldiner, M. et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134, 634–645 (2008)
Leipe, D. D., Wolf, Y. I., Koonin, E. V. & Aravind, L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41–72 (2002)
Zhou, T., Radaev, S., Rosen, B. P. & Gatti, D. L. Structure of the ArsA ATPase: the catalytic subunit of a heavy metal resistance pump. EMBO J. 19, 4838–4845 (2000)
Georgiadis, M. M. et al. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii . Science 257, 1653–1659 (1992)
Leonard, T. A., Butler, P. J. & Lowe, J. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer—a conserved biological switch. EMBO J. 24, 270–282 (2005)
Hayashi, I., Oyama, T. & Morikawa, K. Structural and functional studies of MinD ATPase: implications for the molecular recognition of the bacterial cell division apparatus. EMBO J. 20, 1819–1828 (2001)
Freymann, D. M., Keenan, R. J., Stroud, R. M. & Walter, P. Structure of the conserved GTPase domain of the signal recognition particle. Nature 385, 361–364 (1997)
Montoya, G., Svensson, C., Luirink, J. & Sinning, I. Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature 385, 365–368 (1997)
Metz, J., Wachter, A., Schmidt, B., Bujnicki, J. M. & Schwappach, B. The yeast Arr4p ATPase binds the chloride transporter Gef1p when copper is available in the cytosol. J. Biol. Chem. 281, 410–417 (2006)
Keenan, R. J., Freymann, D. M., Walter, P. & Stroud, R. M. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell 94, 181–191 (1998)
Jiang, Y. et al. Nonequivalence of the nucleotide binding domains of the ArsA ATPase. J. Biol. Chem. 280, 9921–9926 (2005)
Schindelin, H., Kisker, C., Schlessman, J. L., Howard, J. B. & Rees, D. C. Structure of ADP·AIF4 --stabilized nitrogenase complex and its implications for signal transduction. Nature 387, 370–376 (1997)
Sprang, S. R. G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem. 66, 639–678 (1997)
Gasper, R., Meyer, S., Gotthardt, K., Sirajuddin, M. & Wittinghofer, A. It takes two to tango: regulation of G proteins by dimerization. Nature Rev. Mol. Cell Biol. 10, 423–429 (2009)
Zhou, T., Radaev, S., Rosen, B. P. & Gatti, D. L. Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation. J. Biol. Chem. 276, 30414–30422 (2001)
Bernstein, H. D. et al. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature 340, 482–486 (1989)
Horie, C., Suzuki, H., Sakaguchi, M. & Mihara, K. Characterization of signal that directs C-tail-anchored proteins to mammalian mitochondrial outer membrane. Mol. Biol. Cell 13, 1615–1625 (2002)
Kaufmann, T. et al. Characterization of the signal that directs Bcl-xL, but not Bcl-2, to the mitochondrial outer membrane. J. Cell Biol. 160, 53–64 (2003)
Kuroda, R. et al. Charged amino acids at the carboxyl-terminal portions determine the intracellular locations of two isoforms of cytochrome b5. J. Biol. Chem. 273, 31097–31102 (1998)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D. & Winn, M. D. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Kiianitsa, K., Solinger, J. A. & Heyer, W. D. NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high specific activities. Anal. Biochem. 321, 266–271 (2003)
Vogel, G. & Steinhart, R. ATPase of Escherichia coli: purification, dissociation, and reconstitution of the active complex from the isolated subunits. Biochemistry 15, 208–216 (1976)
Van den Berg, B. et al. X-ray structure of a protein-conducting channel. Nature 427, 36–44 (2004)
Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 (1993)
Delano, W. L. The PyMOL Molecular Graphics System 〈http://www.pymol.org〉 (2002)
Acknowledgements
Data were collected at beamlines 21-IDG and 23-IDD at the Advanced Photon Source (APS), Argonne National Laboratory, and we thank the beamline staff for support. Use of the APS was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357. We thank B. Glick and R. Strack for reagents and advice, X. Li for assay characterization, and A. Shiau and T. Steck for comments on the manuscript. This work was supported by a grant from the Edward Mallinckrodt, Jr. Foundation (to R.J.K.) and by the Intramural Research Program of the National Institutes of Health (to R.S.H.).
Author Contributions A.M., A.S. and M.E.D. carried out cloning, protein purification and crystallization. A.M. performed the mutagenesis. M.M. and R.S.H. performed the TA substrate-binding assay. M.D. carried out the ATPase assay. M.E.D. performed the yeast genetic complementation assay. A.M., A.S. and R.J.K. carried out data collection. A.M. and R.J.K. carried out structure determination and model building. R.J.K. supervised the work and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-5 with Legends, Supplementary Tables 1-2 and Supplementary References. (PDF 7170 kb)
Rights and permissions
About this article
Cite this article
Mateja, A., Szlachcic, A., Downing, M. et al. The structural basis of tail-anchored membrane protein recognition by Get3. Nature 461, 361–366 (2009). https://doi.org/10.1038/nature08319
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08319
This article is cited by
-
Mechanism of an intramembrane chaperone for multipass membrane proteins
Nature (2022)
-
Structural and molecular mechanisms for membrane protein biogenesis by the Oxa1 superfamily
Nature Structural & Molecular Biology (2021)
-
Alternative redox forms of ASNA-1 separate insulin signaling from tail-anchored protein targeting and cisplatin resistance in C. elegans
Scientific Reports (2021)
-
The Ways of Tails: the GET Pathway and more
The Protein Journal (2019)
-
In search of tail-anchored protein machinery in plants: reevaluating the role of arsenite transporters
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.