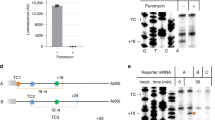
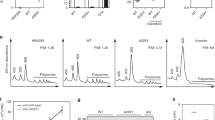
Abstract
The rates of RNA decay and transcription determine the steady-state levels of all messenger RNA and both can be subject to regulation. Although the details of transcriptional regulation are becoming increasingly understood, the mechanism(s) controlling mRNA decay remain unclear. In yeast, a major pathway of mRNA decay begins with deadenylation followed by decapping and 5′–3′ exonuclease digestion. Importantly, it is hypothesized that ribosomes must be removed from mRNA before transcripts are destroyed. Contrary to this prediction, here we show that decay takes place while mRNAs are associated with actively translating ribosomes. The data indicate that dissociation of ribosomes from mRNA is not a prerequisite for decay and we suggest that the 5′–3′ polarity of mRNA degradation has evolved to ensure that the last translocating ribosome can complete translation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Franks, T. M. & Lykke-Andersen, J. The control of mRNA decapping and P-body formation. Mol. Cell 32, 605–615 (2008)
Parker, R. & Sheth, U. P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635–646 (2007)
Coller, J. & Parker, R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73, 861–890 (2004)
Eulalio, A., Behm-Ansmant, I. & Izaurralde, E. P bodies: at the crossroads of post-transcriptional pathways. Nature Rev. Mol. Cell Biol. 8, 9–22 (2007)
Coller, J. & Parker, R. General translational repression by activators of mRNA decapping. Cell 122, 875–886 (2005)
Chu, C. Y. & Rana, T. M. Translation Repression in Human Cells by MicroRNA-Induced Gene Silencing Requires RCK/p54. PLoS Biol. 4, e210 (2006)
He, F. & Jacobson, A. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol. Cell. Biol. 21, 1515–1530 (2001)
Hsu, C. L. & Stevens, A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13, 4826–4835 (1993)
Muhlrad, D., Decker, C. J. & Parker, R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 15, 2145–2156 (1995)
Inada, T. et al. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA 8, 948–958 (2002)
Couttet, P., Fromont-Racine, M., Steel, D., Pictet, R. & Grange, T. Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc. Natl Acad. Sci. USA 94, 5628–5633 (1997)
Amrani, N., Ghosh, S., Mangus, D. A. & Jacobson, A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature 453, 1276–1280 (2008)
Smith, D. W. & McNamara, A. L. Specialization of rabbit reticulocyte transfer RNA content for hemoglobin synthesis. Science 971, 577–579 (1971)
Stevens, A. 5′-exoribonuclease 1: Xrn1. Methods Enzymol. 342, 251–259 (2001)
Doma, M. K. & Parker, R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440, 561–564 (2006)
Jacobson, A. & Peltz, S. W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem. 65, 693–739 (1996)
Moore, M. J. & Query, C. C. Joining of RNAs by splinted ligation. Methods Enzymol. 317, 109–123 (2000)
Beelman, C. A. & Parker, R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J. Biol. Chem. 269, 9687–9692 (1994)
Mangus, D. A. & Jacobson, A. Linking mRNA turnover and translation: assessing the polyribosomal association of mRNA decay factors and degradative intermediates. Methods 17, 28–37 (1999)
Decker, C. J., Teixeira, D. & Parker, R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae . J. Cell Biol. 179, 437–449 (2007)
Sweet, T. J., Boyer, B., Hu, W., Baker, K. E. & Coller, J. Microtubule disruption stimulates P-body formation. RNA 13, 493–502 (2007)
Eulalio, A., Behm-Ansmant, I., Schweizer, D. & Izaurralde, E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 27, 3970–3981 (2007)
Wang, Z., Jiao, X., Carr-Schmid, A. & Kiledjian, M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl Acad. Sci. USA 99, 12663–12668 (2002)
Acknowledgements
We thank P. Maroney and T. Nilsen for sharing unpublished data about the poly(A) tailing assay. We also thank T. Nilsen for his insight, suggestions and evaluation of the manuscript. We also thank R. Parker, A. van Hoof and M. Wickens for reagents and advice. Funding was provided by the American Heart Association and the National Institutes of Health.
Author Contributions W.H., K.E.B. and J.C. wrote the manuscript. W.H. and T.J.S. performed the experiments. S.C. provided technical expertise with poly(A) tailing assays. All of the authors contributed to discussion and the design of the research. All authors commented on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Notes, Supplementary Figures S1-S10 with Legends and Supplementary Tables 1-2. (PDF 2801 kb)
Rights and permissions
About this article
Cite this article
Hu, W., Sweet, T., Chamnongpol, S. et al. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 461, 225–229 (2009). https://doi.org/10.1038/nature08265
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08265
This article is cited by
-
ADAR1-mediated RNA editing of SCD1 drives drug resistance and self-renewal in gastric cancer
Nature Communications (2023)
-
Observation of conformational changes that underlie the catalytic cycle of Xrn2
Nature Chemical Biology (2022)
-
The effects of codon bias and optimality on mRNA and protein regulation
Cellular and Molecular Life Sciences (2021)
-
Quantification of mRNA translation in live cells using single-molecule imaging
Nature Protocols (2020)
-
Quantification of translation uncovers the functions of the alternative transcriptome
Nature Structural & Molecular Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.