Abstract
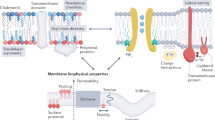
Studies of membrane proteins have revealed a direct link between the lipid environment and the structure and function of some of these proteins. Although some of these effects involve specific chemical interactions between lipids and protein residues, many can be understood in terms of protein-induced perturbations to the membrane shape. The free-energy cost of such perturbations can be estimated quantitatively, and measurements of channel gating in model systems of membrane proteins with their lipid partners are now confirming predictions of simple models.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Engelman, D. M. Membranes are more mosaic than fluid. Nature 438, 578–580 (2005).
Sukharev, S. I. et al. Energetic and spatial parameters for gating of the bacterial large conductance mechanosensitive channel, MscL. J. Gen. Physiol. 113, 525–540 (1999).
Perozo, E. et al. Physical principles underlying the transduction of bilayer deformation forces during mechanosensitive channel gating. Nature Struct. Biol. 9, 696–703 (2002). This paper describes experiments that show how the gating behaviour of mechanosensitive channels depends on the acyl chain lengths of the lipids around them.
Andersen, O.S. & Koeppe, R.E. II Bilayer thickness and membrane protein function: an energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 36, 107–130 (2007). This useful review provides a variety of examples of the connection between the lipid environment and the function of membrane-bound proteins.
Marsh, D. Protein modulation of lipids, and vice-versa, in membranes. Biochim. Biophys. Acta 1778, 1545–1575 (2008).
Killian, J. A. Hydrophobic mismatch between proteins and lipids in membranes. Biochim. Biophys. Acta 1376, 401–415 (1998).
Petsko, G. & Ringe, D. Protein Structure and Function (New Science Press, 2004).
Moe, P. & Blount, P. Assessment of potential stimuli for mechano-dependent gating of MscL: effects of pressure, tension, and lipid headgroups. Biochemistry 44, 12239–12244 (2005).
Lee, A. G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta 1666, 62–87 (2004). This review discusses how specific residues in the structure of transmembrane proteins determine hydrophobic thickness and addresses the topic of site-specific lipid binding.
Valiyaveetil, F. I., Zhou, Y. & MacKinnon, R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry 41, 10771–10777 (2002).
Lundbaek, J. A. et al. Capsaicin regulates voltage-dependent sodium channels by altering lipid bilayer elasticity. Mol. Pharmacol. 68, 680–689 (2005).
Suchyna, T. M. et al. Bilayer-dependent inhibition of mechanosensitive channels by neuroactive peptide enantiomers. Nature 430, 235–240 (2004).
Jensen, M. O. & Mouritsen, O. G. Lipids do influence protein function — the hydrophobic matching hypothesis revisited. Biochim. Biophys. Acta 1666, 205–226 (2004). This review discusses the qualitative effects of thickness mismatch on membrane proteins and summarizes evidence of the effects of bilayer thickness on enzymatic activity.
Goulian, M. et al. Gramicidin channel kinetics under tension. Biophys. J. 74, 328–337 (1998).
Schmidt, D. & MacKinnon, R. Voltage-dependent K+ channel gating and voltage sensor toxin sensitivity depend on the mechanical state of the lipid membrane. Proc. Natl Acad. Sci. USA 105, 19276–19281 (2008).
Calabrese, B. et al. Mechanosensitivity of N-type calcium channel currents. Biophys. J. 83, 2560–2574 (2002).
Morris, C. E. & Juranka, P. F. Nav channel mechanosensitivity: activation and inactivation accelerate reversibly with stretch. Biophys. J. 93, 822–833 (2007).
Takamori, S. et al. Molecular anatomy of a trafficking organelle. Cell 127, 831–846 (2006). This paper provides an experimental treatment of the crowded nature of biological membranes.
Dupuy, A. D. & Engelman, D. M. Protein area occupancy at the center of the red blood cell membrane. Proc. Natl Acad. Sci. USA 105, 2848–2852 (2008).
Mitra, K. et al. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc. Natl Acad. Sci. USA 101, 4083–4088 (2004). This experimental tour de force shows that protein content modulates bilayer thickness across different organellar membranes by the hydrophobic mismatch principle.
Zimmerman, S. B. & Minton, A. P. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 22, 27–65 (1993).
Ellis, R. J. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 26, 597–604 (2001).
Bond, P. J. & Sansom, M. S. Bilayer deformation by the Kv channel voltage sensor domain revealed by self-assembly simulations. Proc. Natl Acad. Sci. USA 104, 2631–2636 (2007).
Chen, X. et al. Gating mechanisms of mechanosensitive channels of large conductance, I: a continuum mechanics-based hierarchical framework. Biophys. J. 95, 563–580 (2008).
Elmore, D. E. & Dougherty, D. A. Investigating lipid composition effects on the mechanosensitive channel of large conductance (MscL) using molecular dynamics simulations. Biophys. J. 85, 1512–1524 (2003).
Jeon, J. & Voth, G. A. Gating of the mechanosensitive channel protein MscL: the interplay of membrane and protein. Biophys. J. 94, 3497–3511 (2008).
Marsh, D. Lateral pressure profile, spontaneous curvature frustration, and the incorporation and conformation of proteins in membranes. Biophys. J. 93, 3884–3899 (2007).
Cantor, R. The influence of membrane lateral pressures on simple geometric models of protein conformational equilibria. Chem. Phys. Lipids 101, 45–56 (1999).
Marsh, D. Elastic curvature constants of lipid monolayers and bilayers. Chem. Phys. Lipids 144, 146–159 (2006). This review compiles measurements of membrane elastic properties across different lipid types and builds scaling laws that connect different modes of bilayer deformation to bulk bilayer properties.
Dan, N., Pincus, P. & Safran, S. Membrane-induced interactions between inclusions. Langmuir 9, 2768–2771 (1993).
Markin, V. S. & Sachs, F. Thermodynamics of mechanosensitivity. Phys. Biol. 1, 110–124 (2004).
Mouritsen, O. G. & Bloom, M. Models of lipid-protein interactions in membranes. Annu. Rev. Biophys. Biomol. Struct. 22, 145–171 (1993).
Boal, D. Mechanics of the Cell (Cambridge University Press, 2002).
Gruner, S. M. Intrinsic curvature hypothesis for biomembrane lipid composition: a role for nonbilayer lipids. Proc. Natl Acad. Sci. USA 82, 3665–3669 (1985).
Mouritsen, O. G. & Bloom, M. Mattress model of lipid–protein interactions in membranes. Biophys. J. 46, 141–153 (1984).
Huang, H. W., Deformation free energy of bilayer membrane and its effect on gramicidin channel lifetime. Biophys. J. 50, 1061–1070 (1986).
Turner, M. S. & Sens, P. Gating-by-tilt of mechanically sensitive membrane channels. Phys. Rev. Lett. 93, 118103 (2004).
Wiggins, P. & Phillips, R. Membrane-protein interactions in mechanosensitive channels. Biophys. J. 88, 880–902 (2005).
Turner, M. S. & Sens, P. Inclusions on fluid membranes anchored to elastic media. Biophys. J. 76, 564–572 (1999).
Wiggins, P. & Phillips, R. Analytic models for mechanotransduction: gating a mechanosensitive channel. Proc. Natl Acad. Sci. USA 101, 4071–4076 (2004).
Dan, N. & Safran, S. A. Effect of lipid characteristics on the structure of transmembrane proteins. Biophys. J. 75, 1410–1414 (1998).
Nielsen, C., Goulian, M. & Andersen, O. S. Energetics of inclusion-induced bilayer deformations. Biophys. J. 74, 1966–1983 (1998).
Ursell, T., Reeves, D., Wiggins, P. & Phillips, R. in Mechanosensitive Ion Channels (ed. Kamkin, A. & Kisileva, I.) 37–70 (Springer, 2008).
Cantor, R. S. Lateral pressures in cell membranes: A mechanism for modulation of protein function. J. Phys. Chem. B 101, 1723–1725 (1997).
Bruinsma, R., Goulian, M. & Pincus, P. Self-assembly of membrane junctions. Biophys. J. 67, 746–750 (1994).
Gil, T. et al. Theoretical analysis of protein organization in lipid membranes. Biochim. Biophys. Acta 1376, 245–266 (1998).
Muller, M. M., Deserno, M. & Guven, J. Interface-mediated interactions between particles: a geometrical approach. Phys. Rev. E 72, 061407 (2005).
Reynwar, B. J. et al. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature 447, 461–464 (2007).
Golestanian, R., Goulian, M. & Kardar, M. Fluctuation-induced interactions between rods on a membrane. Phys. Rev. E 54, 6725–6734 (1996).
Goulian, M., Bruinsma, R. & Pincus, P. Long-range forces in heterogeneous fluid membranes. Europhys. Lett. 22, 145–150 (1993).
Sheetz, M.P., Sable, J.E. & Dobereiner, H. G. Continuous membrane-cytoskeleton adhesion requires continuous accommodation to lipid and cytoskeleton dynamics. Annu. Rev. Biophys. Biomol. Struct. 35, 417–434 (2006).
Weikl, T. R., Kozlov, M. M. & Helfrich, W. Interaction of conical membrane inclusions: Effect of lateral tension. Phys. Rev. E 57, 6988–6995 (1998).
Goforth, R. L. et al. Hydrophobic coupling of lipid bilayer energetics to channel function. J. Gen. Physiol. 121, 477–493 (2003).
Sens, P., Johannes, L. & Bassereau, P. Biophysical approaches to protein-induced membrane deformations in trafficking. Curr. Opin. Cell Biol. 20, 476–482 (2008).
Blood, P. D. & Voth, G. A. Direct observation of Bin/amphiphysin/Rvs (BAR) domain-induced membrane curvature by means of molecular dynamics simulations. Proc. Natl Acad. Sci. USA 103, 15068–15072 (2006).
Reynwar, B. J. et al. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature 447, 461–464 (2007).
Arkhipov, A., Yin, Y. & Schulten, K. Four-scale description of membrane sculpting by BAR domains. Biophys. J. 95, 2806–2821 (2008).
Kim, K. S., Neu, J. & Oster, G. Curvature-mediated interactions between membrane proteins. Biophys. J. 75, 2274–2291 (1998).
Leibler, S. Curvature instability in membranes. J. Physique 47, 507–516 (1986).
Sens, P. & Turner, M. S. Theoretical model for the formation of caveolae and similar membrane invaginations. Biophys. J. 86, 2049–2057 (2004).
Ursell, T. et al. Cooperative gating and spatial organization of membrane proteins through elastic interactions. PLoS Comput. Biol. 3, e81 (2007).
Zhou, H. X., Rivas, G. & Minton, A. P. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 37, 375–397 (2008).
Chang, G. et al. Structure of the MscL homolog from Mycobacterium tuberculosis: a gated mechanosensitive ion channel. Science 282, 2220–2226 (1998).
Bass, R. B. et al. Crystal structure of Escherichia coli MSCS, a voltage-modulated and mechanosensitive channel. Science 298, 1582–1587 (2002).
Acknowledgements
We are grateful to many people for insights and comments. In particular, we thank O. Andersen, M. Bialecka, F. Brown, N. Dan, L. Haswell, S. Johnson, J. Kondev, R. MacKinnon, C. Morris, D. Rees, D. Reeves, F. Sachs, D. Schmidt, S. Scheuring, S. Sukharev, M. Turner, S. White and M. Widom. We are also grateful to several anonymous reviewers for insightful comments. In addition, we are grateful to the US National Science Foundation and the National Institutes of Health for support through NIH Award number R01 GM084211 and the Director's Pioneer Award. The reference list for this article was constrained by length limits, and as a result the references cited here are representative rather than comprehensive. We apologize to those whose references are not cited as a result of either space limitations or our ignorance.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reprints and permissions information is available at http://www.nature.com/reprints.
Correspondence should be addressed to R.P. (phillips@pboc.caltech.edu).
Rights and permissions
About this article
Cite this article
Phillips, R., Ursell, T., Wiggins, P. et al. Emerging roles for lipids in shaping membrane-protein function. Nature 459, 379–385 (2009). https://doi.org/10.1038/nature08147
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08147
This article is cited by
-
Photolipid excitation triggers depolarizing optocapacitive currents and action potentials
Nature Communications (2024)
-
Spiral packing and chiral selectivity in model membranes probed by phase-resolved sum-frequency generation microscopy
Nature Communications (2024)
-
Structural heterogeneity of the ion and lipid channel TMEM16F
Nature Communications (2024)
-
A concerted ATPase cycle of the protein transporter AAA-ATPase Bcs1
Nature Communications (2023)
-
Photoinduced bidirectional switching in lipid membranes containing azobenzene glycolipids
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.