Abstract
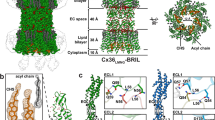
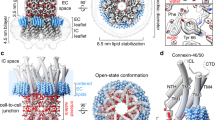
Gap junctions consist of arrays of intercellular channels between adjacent cells that permit the exchange of ions and small molecules. Here we report the crystal structure of the gap junction channel formed by human connexin 26 (Cx26, also known as GJB2) at 3.5 Å resolution, and discuss structural determinants of solute transport through the channel. The density map showed the two membrane-spanning hemichannels and the arrangement of the four transmembrane helices of the six protomers forming each hemichannel. The hemichannels feature a positively charged cytoplasmic entrance, a funnel, a negatively charged transmembrane pathway, and an extracellular cavity. The pore is narrowed at the funnel, which is formed by the six amino-terminal helices lining the wall of the channel, which thus determines the molecular size restriction at the channel entrance. The structure of the Cx26 gap junction channel also has implications for the gating of the channel by the transjunctional voltage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kumar, N. M. & Gilula, N. B. The gap junction communication channel. Cell 84, 381–388 (1996)
Harris, A. L. Emerging issues of connexin channels: biophysics fills the gap. Q. Rev. Biophys. 34, 325–472 (2001)
Foote, C. I., Zhou, L., Zhu, X. & Nicholson, B. J. The pattern of disulfide linkages in the extracellular loop regions of connexin 32 suggests a model for the docking interface of gap junctions. J. Cell Biol. 140, 1187–1197 (1998)
Levin, M. Gap junctional communication in morphogenesis. Prog. Biophys. Mol. Biol. 94, 186–206 (2007)
Saez, J. C., Berthoud, V. M., Branes, M. C., Martinez, A. D. & Beyer, E. C. Plasma membrane channels formed by connexins: their regulation and functions. Physiol. Rev. 83, 1359–1400 (2003)
Kelsell, D. P., Dunlop, J. & Hodgins, M. B. Human diseases: clues to cracking the connexin code? Trends Cell Biol. 11, 2–6 (2001)
Simon, A. M. & Goodenough, D. A. Diverse functions of vertebrate gap junctions. Trends Cell Biol. 8, 477–483 (1998)
Unwin, P. N. & Zampighi, G. Structure of the junction between communicating cells. Nature 283, 545–549 (1980)
Unwin, P. N. & Ennis, P. D. Two configurations of a channel-forming membrane protein. Nature 307, 609–613 (1984)
Unger, V. M., Kumar, N. M., Gilula, N. B. & Yeager, M. Three-dimensional structure of a recombinant gap junction membrane channel. Science 283, 1176–1180 (1999)
Fleishman, S. J., Unger, V. M., Yeager, M. & Ben-Tal, N. A C-α model for the transmembrane α helices of gap junction intercellular channels. Mol. Cell 15, 879–888 (2004)
Oshima, A., Tani, K., Hiroaki, Y., Fujiyoshi, Y. & Sosinsky, G. E. Three-dimensional structure of a human connexin26 gap junction channel reveals a plug in the vestibule. Proc. Natl Acad. Sci. USA 104, 10034–10039 (2007)
Harris, A. L., Spray, D. C. & Bennett, M. V. Kinetic properties of a voltage-dependent junctional conductance. J. Gen. Physiol. 77, 95–117 (1981)
Verselis, V. K., Ginter, C. S. & Bargiello, T. A. Opposite voltage gating polarities of two closely related connexins. Nature 368, 348–351 (1994)
Ebihara, L., Berthoud, V. M. & Beyer, E. C. Distinct behavior of connexin56 and connexin46 gap junctional channels can be predicted from the behavior of their hemi-gap-junctional channels. Biophys. J. 68, 1796–1803 (1995)
Bukauskas, F. F., Bukauskiene, A., Bennett, M. V. & Verselis, V. K. Gating properties of gap junction channels assembled from connexin 43 and connexin 43 fused with green fluorescent protein. Biophys. J. 81, 137–152 (2001)
Bukauskas, F. F. & Verselis, V. K. Gap junction channel gating. Biochim. Biophys. Acta 1662, 42–60 (2004)
Muller, D. J., Hand, G. M., Engel, A. & Sosinsky, G. E. Conformational changes in surface structures of isolated connexin 26 gap junctions. EMBO J. 21, 3598–3607 (2002)
Perkins, G. A., Goodenough, D. A. & Sosinsky, G. E. Formation of the gap junction intercellular channel requires a 30° rotation for interdigitating two apposing connexons. J. Mol. Biol. 277, 171–177 (1998)
Skerrett, I. M. et al. Identification of amimo acid residues lining the pore of a gap junction channel. J. Cell Biol. 159, 349–360 (2002)
Zhou, X. W. et al. Identification of a pore lining segment in gap junction hemichannels. Biophys. J. 72, 1946–1953 (1997)
Kronengold, J., Trexler, E. B., Bukauskas, F. F., Bargiello, T. A. & Verselis, V. K. Single-channel SCAM identifies pore-lining residues in the first extracellular loop and first transmembrane domains of Cx46 hemichannels. J. Gen. Physiol. 122, 389–405 (2003)
Suchyna, T. M., Xu, L. X., Gao, F., Fourtner, C. R. & Nicholson, B. J. Identification of a proline residue as a transduction element involved in voltage gating of gap junctions. Nature 365, 847–849 (1993)
Laird, D. W. Life cycle of connexins in health and disease. Biochem. J. 394, 527–543 (2006)
Sheridan, R. P., Levy, R. M. & Salemme, F. R. α-helix dipole model and electrostatic stabilization of 4-α-helical proteins. Proc. Natl Acad. Sci. USA 79, 4545–4549 (1982)
Weber, P. A., Chang, H. C., Spaeth, K. E., Nitsche, J. M. & Nicholson, B. J. The permeability of gap junction channels to probes of different size is dependent on connexin composition and permeant-pore affinities. Biophys. J. 87, 958–973 (2004)
Oh, S., Verselis, V. K. & Bargiello, T. A. Charges dispersed over the permeation pathway determine the charge selectivity and conductance of a Cx32 chimeric hemichannel. J. Physiol. (Lond.) 586, 2445–2461 (2008)
Gong, X. Q. & Nicholson, B. J. Size selectivity between gap junction channels composed of different connexins. Cell Commun. Adhes. 8, 187–192 (2001)
Trexler, E. B., Bukauskas, F. F., Kronengold, J., Bargiello, T. A. & Verselis, V. K. The first extracellular loop domain is a major determinant of charge selectivity in connexin46 channels. Biophys. J. 79, 3036–3051 (2000)
Purnick, P. E., Benjamin, D. C., Verselis, V. K., Bargiello, T. A. & Dowd, T. L. Structure of the amino terminus of a gap junction protein. Arch. Biochem. Biophys. 381, 181–190 (2000)
Miyazawa, A., Fujiyoshi, Y. & Unwin, N. Structure and gating mechanism of the acetylcholine receptor pore. Nature 423, 949–955 (2003)
Kelsell, D. P. et al. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature 387, 80–83 (1997)
Oshima, A., Doi, T., Mitsuoka, K., Maeda, S. & Fujiyoshi, Y. Roles of Met-34, Cys-64, and Arg-75 in the assembly of human connexin 26. Implication for key amino acid residues for channel formation and function. J. Biol. Chem. 278, 1807–1816 (2003)
Oshima, A., Tani, K., Hiroaki, Y., Fujiyoshi, Y. & Sosinsky, G. E. Projection structure of a N-terminal deletion mutant of connexin 26 channel with decreased central pore density. Cell Commun. Adhes. 15, 85–93 (2008)
Purnick, P. E., Oh, S., Abrams, C. K., Verselis, V. K. & Bargiello, T. A. Reversal of the gating polarity of gap junctions by negative charge substitutions in the N-terminus of connexin 32. Biophys. J. 79, 2403–2415 (2000)
Oh, S., Rivkin, S., Tang, Q., Verselis, V. K. & Bargiello, T. A. Determinants of gating polarity of a connexin 32 hemichannel. Biophys. J. 87, 912–928 (2004)
Oh, S., Abrams, C. K., Verselis, V. K. & Bargiello, T. A. Stoichiometry of transjunctional voltage-gating polarity reversal by a negative charge substitution in the amino terminus of a connexin 32 chimera. J. Gen. Physiol. 116, 13–31 (2000)
Jan, L. Y. & Jan, Y. N. Structural elements involved in specific K+ channel functions. Annu. Rev. Physiol. 54, 537–555 (1992)
Trexler, E. B., Bennett, M. V. L., Bargiello, T. A. & Verselis, V. K. Voltage gating and permeation in a gap junction hemichannel. Proc. Natl Acad. Sci. USA 93, 5836–5841 (1996)
Peracchia, C. Chemical gating of gap junction channels; roles of calcium, pH and calmodulin. Biochim. Biophys. Acta 1662, 61–80 (2004)
Delmar, M., Coombs, W., Sorgen, P., Duffy, H. S. & Taffet, S. M. Structural bases for the chemical regulation of connexin43 channels. Cardiovasc. Res. 62, 268–275 (2004)
Tao, L. & Harris, A. L. 2-Aminoethoxydiphenyl borate directly inhibits channels composed of connexin26 and/or connexin32. Mol. Pharmacol. 71, 570–579 (2007)
Baker, N. A., Sept, D., Joseph, S. & Holst, M. J. McCammon. J. A. Electrostatics of nanosystems: applications to microtubles and the ribosomes. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001)
Stauffer, K. A., Kumar, N. M., Gilula, N. B. & Unwin, N. Isolation and purification of gap junction channels. J. Cell Biol. 115, 141–150 (1991)
Bellizzi, J. J., Widom, J., Kemp, C. W. & Clardy, J. Producing selenomethionine-labeled proteins with a baculovirus expression vector system. Structure 7, R263–R267 (1999)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative Computational Project 4 The CCP4 suite: Programs for Protein Crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993)
Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D 59, 2023–2030 (2003)
Cowtan, K. An automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newsletter Protein Crystallogr. 31, 34–38 (1994)
Cowtan, K. D. & Zhang, K. Y. Density modification for macromolecular phase improvement. Prog. Biophys. Mol. Biol. 72, 245–270 (1999)
Jones, T. A., Zou, J. Y. & Cowan, S. W. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Brunger, A. T. et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Murshudov, G. N., Vagin, A. A. & Dadson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Dodson, E., Kleywegt, G. J. & Wilson, K. Report of a workshop on the use of statistical validators in protein X-ray crystallography. Acta. Crystallogr. D 52, 228–234 (1996)
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993)
Delano, W. L. The PyMOL Molecular Graphics System. v.0.99 (Delano Scientific, 2006)
Acknowledgements
We thank T. Tomizaki for help in the diffraction data collection on X06SA at the Swiss Light Source. This work was supported by Grants-in-Aid for Scientific Research (10687101, 16087206 and 18207006) and the GCOE program (A-041) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to T.T.), the Japan Biological Informatics Consortium (to T.T.), the Strategic Japan-UK Cooperation Program of the Japan Science and Technology Agency (to T.T.), and Grants-in-Aid for Specially Promoted Research (to Y.F.) and the New Energy and Industrial Technology Development Organization (to Y.F.). We thank T. Walz for critical reading of this manuscript.
Author Contributions S.M., S.N., M.S., E.Y. and T.T. performed X-ray structural analysis. S.M., A.O., Y.F. and T.T. wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-11 with Legends, Supplementary References, a Supplementary Discussion and Supplementary Table 1 (PDF 3771 kb)
Rights and permissions
About this article
Cite this article
Maeda, S., Nakagawa, S., Suga, M. et al. Structure of the connexin 26 gap junction channel at 3.5 Å resolution. Nature 458, 597–602 (2009). https://doi.org/10.1038/nature07869
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature07869
This article is cited by
-
Association of variants in GJA8 with familial acorea-microphthalmia-cataract syndrome
European Journal of Human Genetics (2023)
-
A Descriptive Observational Study of GJB2 and GJB6 Mutations in Familial Autosomal Recessive Non-syndromic Hearing Impairment
Indian Journal of Otolaryngology and Head & Neck Surgery (2023)
-
Biallelic mutations in pakistani families with autosomal recessive prelingual nonsyndromic hearing loss
Genes & Genomics (2023)
-
Ab initio phasing macromolecular structures using electron-counted MicroED data
Nature Methods (2022)
-
Altered neural cell junctions and ion-channels leading to disrupted neuron communication in Parkinson’s disease
npj Parkinson's Disease (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.