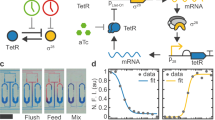
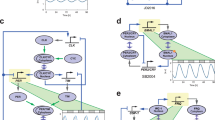
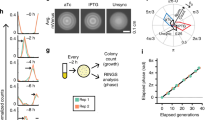
Abstract
Autonomous and self-sustained oscillator circuits mediating the periodic induction of specific target genes are minimal genetic time-keeping devices found in the central and peripheral circadian clocks1,2. They have attracted significant attention because of their intriguing dynamics and their importance in controlling critical repair3, metabolic4 and signalling pathways5. The precise molecular mechanism and expression dynamics of this mammalian circadian clock are still not fully understood. Here we describe a synthetic mammalian oscillator based on an auto-regulated sense–antisense transcription control circuit encoding a positive and a time-delayed negative feedback loop, enabling autonomous, self-sustained and tunable oscillatory gene expression. After detailed systems design with experimental analyses and mathematical modelling, we monitored oscillating concentrations of green fluorescent protein with tunable frequency and amplitude by time-lapse microscopy in real time in individual Chinese hamster ovary cells. The synthetic mammalian clock may provide an insight into the dynamics of natural periodic processes and foster advances in the design of prosthetic networks in future gene and cell therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gillette, M. U. & Sejnowski, T. J. Physiology. Biological clocks coordinately keep life on time. Science 309, 1196–1198 (2005)
Schibler, U. & Sassone-Corsi, P. A web of circadian pacemakers. Cell 111, 919–922 (2002)
Lahav, G. The strength of indecisiveness: oscillatory behavior for better cell fate determination. Sci. STKE 2004, pe55 (2004)
Kaasik, K. & Lee, C. C. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 430, 467–471 (2004)
Covert, M. W., Leung, T. H., Gaston, J. E. & Baltimore, D. Achieving stability of lipopolysaccharide-induced NF-κB activation. Science 309, 1854–1857 (2005)
Hasty, J., McMillen, D. & Collins, J. J. Engineered gene circuits. Nature 420, 224–230 (2002)
Reppert, S. M. & Weaver, D. R. Forward genetic approach strikes gold: cloning of a mammalian clock gene. Cell 89, 487–490 (1997)
Storch, K. F. et al. Extensive and divergent circadian gene expression in liver and heart. Nature 417, 78–83 (2002)
Reppert, S. M. & Weaver, D. R. Coordination of circadian timing in mammals. Nature 418, 935–941 (2002)
Nagoshi, E. et al. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119, 693–705 (2004)
Gekakis, N. et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569 (1998)
Atkinson, M. R., Savageau, M. A., Myers, J. T. & Ninfa, A. J. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli . Cell 113, 597–607 (2003)
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000)
Fung, E. et al. A synthetic gene–metabolic oscillator. Nature 435, 118–122 (2005)
Chilov, D. & Fussenegger, M. Toward construction of a self-sustained clock-like expression system based on the mammalian circadian clock. Biotechnol. Bioeng. 87, 234–242 (2004)
Gossen, M. & Bujard, H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA 89, 5547–5551 (1992)
Fussenegger, M. et al. Streptogramin-based gene regulation systems for mammalian cells. Nature Biotechnol. 18, 1203–1208 (2000)
Kramer, B. P. & Fussenegger, M. Transgene control engineering in mammalian cells. Methods Mol. Biol. 308, 123–143 (2005)
Rossmanith, W., Chabicovsky, M., Herkner, K. & Schulte-Hermann, R. Cellular gene dose and kinetics of gene expression in mouse livers transfected by high-volume tail-vein injection of naked DNA. DNA Cell Biol. 21, 847–853 (2002)
Rosenfeld, N., Young, J. W., Alon, U., Swain, P. S. & Elowitz, M. B. Gene regulation at the single-cell level. Science 307, 1962–1965 (2005)
Vilar, J. M., Kueh, H. Y., Barkai, N. & Leibler, S. Mechanisms of noise-resistance in genetic oscillators. Proc. Natl Acad. Sci. USA 99, 5988–5992 (2002)
Di Ventura, B., Lemerle, C., Michalodimitrakis, K. & Serrano, L. From in vivo to in silico biology and back. Nature 443, 527–533 (2006)
Crosthwaite, S. K. Circadian clocks and natural antisense RNA. FEBS Lett. 567, 49–54 (2004)
Morton, A. J. et al. Disintegration of the sleep–wake cycle and circadian timing in Huntingdon’s disease. J. Neurosci. 25, 157–163 (2005)
Wu, Y. H. et al. Pineal clock gene oscillation is disturbed in Alzheimer’s disease, due to functional disconnection from the ‘master clock’. FASEB J. 20, 1874–1876 (2006)
Johnson, E. S., Ma, P. C., Ota, I. M. & Varshavsky, A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270, 17442–17456 (1995)
Fux, C. et al. Streptogramin- and tetracycline-responsive dual regulated expression of p27Kip1 sense and antisense enables positive and negative growth control of Chinese hamster ovary cells. Nucleic Acids Res. 29, e19 (2001)
Moser, S. et al. Dual-regulated expression technology: a new era in the adjustment of heterologous gene expression in mammalian cells. J. Gene Med. 3, 529–549 (2001)
Fussenegger, M., Schlatter, S., Datwyler, D., Mazur, X. & Bailey, J. E. Controlled proliferation by multigene metabolic engineering enhances the productivity of Chinese hamster ovary cells. Nature Biotechnol. 16, 468–472 (1998)
Acknowledgements
We thank H. Meyer for providing pcDNA3.1-UbV76–GFP; and B. Kramer, M. Gitzinger, D. Greber and W. Weber for conceptual input and/or critical comments on the manuscript. This work was supported by the Swiss National Science Foundation and the EC Framework 6 (COBIOS).
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
This file contains Supplementary Notes and Methods, Supplementary References, Supplementary Tables 1-5, and Supplementary Figures 1-9 with Legends (PDF 2632 kb)
Supplementary Movie
Supplementary Movie 1 shows that CHO-K1 were co-transfected at equimolar ratios (100ng each) with pMT35 (PhCMV*-1⇐tTA←PPIR), pMT36 (PhCMV*-1⇐PIT) and pMT100 (PhCMV*-1⇐UbV76-GFP) and cultivated in the absence of antibiotics. Several CHO-K1 cells show oscillating UbV76-GFP expression. The fluorescence diagrams of the cells which are surrounded by circles are shown in Fig. 3. (MOV 6432 kb)
Rights and permissions
About this article
Cite this article
Tigges, M., Marquez-Lago, T., Stelling, J. et al. A tunable synthetic mammalian oscillator. Nature 457, 309–312 (2009). https://doi.org/10.1038/nature07616
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature07616
This article is cited by
-
Phenotype Design Space Provides a Mechanistic Framework Relating Molecular Parameters to Phenotype Diversity Available for Selection
Journal of Molecular Evolution (2023)
-
Complex dynamics in a synchronized cell-free genetic clock
Nature Communications (2022)
-
Predicting synchronized gene coexpression patterns from fibration symmetries in gene regulatory networks in bacteria
BMC Bioinformatics (2021)
-
Towards a physical understanding of developmental patterning
Nature Reviews Genetics (2021)
-
Towards organism-level systems biology by next-generation genetics and whole-organ cell profiling
Biophysical Reviews (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.