Abstract
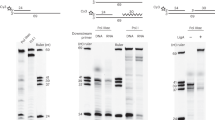
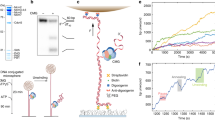
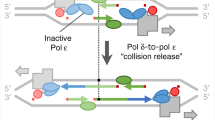
In all organisms, the protein machinery responsible for the replication of DNA, the replisome, is faced with a directionality problem. The antiparallel nature of duplex DNA permits the leading-strand polymerase to advance in a continuous fashion, but forces the lagging-strand polymerase to synthesize in the opposite direction. By extending RNA primers, the lagging-strand polymerase restarts at short intervals and produces Okazaki fragments1,2. At least in prokaryotic systems, this directionality problem is solved by the formation of a loop in the lagging strand of the replication fork to reorient the lagging-strand DNA polymerase so that it advances in parallel with the leading-strand polymerase. The replication loop grows and shrinks during each cycle of Okazaki fragment synthesis3. Here we use single-molecule techniques to visualize, in real time, the formation and release of replication loops by individual replisomes of bacteriophage T7 supporting coordinated DNA replication. Analysis of the distributions of loop sizes and lag times between loops reveals that initiation of primer synthesis and the completion of an Okazaki fragment each serve as a trigger for loop release. The presence of two triggers may represent a fail-safe mechanism ensuring the timely reset of the replisome after the synthesis of every Okazaki fragment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Benkovic, S. J., Valentine, A. M. & Salinas, F. Replisome-mediated DNA replication. Annu. Rev. Biochem. 70, 181–208 (2001)
Johnson, A. & O’Donnell, M. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 74, 283–315 (2005)
Alberts, B. M. et al. Studies on DNA replication in the bacteriophage T4 in vitro system. Cold Spring Harb. Symp. Quant. Biol. 47, 655–668 (1983)
Chastain, P. D., Makhov, A. M., Nossal, N. G. & Griffith, J. D. Analysis of the Okazaki fragment distributions along single long DNAs replicated by the bacteriophage T4 proteins. Mol. Cell 6, 803–814 (2000)
Park, K., Debyser, Z., Tabor, S., Richardson, C. C. & Griffith, J. D. Formation of a DNA loop at the replication fork generated by bacteriophage T7 replication proteins. J. Biol. Chem. 273, 5260–5270 (1998)
Carver, T. E., Sexton, D. J. & Benkovic, S. J. Dissociation of bacteriophage T4 DNA polymerase and its processivity clamp after completion of Okazaki fragment synthesis. Biochemistry 36, 14409–14417 (1997)
López de Saro, F. J., Georgescu, R. E. & O’Donnell, M. A peptide switch regulates DNA polymerase processivity. Proc. Natl Acad. Sci. USA 100, 14689–14694 (2003)
Hacker, K. J. & Alberts, B. M. The rapid dissociation of the T4 DNA-polymerase holoenzyme when stopped by a DNA hairpin helix: a model for polymerase release following the termination of each Okazaki fragment. J. Biol. Chem. 269, 24221–24228 (1994)
Li, X. & Marians, K. J. Two distinct triggers for cycling of the lagging strand polymerase at the replication fork. J. Biol. Chem. 275, 34757–34765 (2000)
Yang, J., Nelson, S. W. & Benkovic, S. J. The control mechanism for lagging strand polymerase recycling during bacteriophage T4 DNA replication. Mol. Cell 21, 153–164 (2006)
Nossal, N. G., Makhov, A. M., Chastain, P. D., Jones, C. E. & Griffith, J. D. Architecture of the bacteriophage T4 replication complex revealed with nanoscale biopointers. J. Biol. Chem. 282, 1098–1108 (2007)
Wu, C. A., Zechner, E. L., Reems, J. A., McHenry, C. S. & Marians, K. J. Coordinated leading-strand and lagging-strand synthesis at the Escherichia coli DNA-replication fork: primase action regulates the cycle of Okazaki fragment synthesis. J. Biol. Chem. 267, 4074–4083 (1992)
Lee, J., Chastain, P. D., Griffith, J. D. & Richardson, C. C. Lagging strand synthesis in coordinated DNA synthesis by bacteriophage T7 replication proteins. J. Mol. Biol. 316, 19–34 (2002)
Tougu, K. & Marians, K. J. The interaction between helicase and primase sets the replication fork clock. J. Biol. Chem. 271, 21398–21405 (1996)
Richardson, C. C. Bacteriophage T7: minimal requirements for the replication of a duplex DNA molecule. Cell 33, 315–317 (1983)
Lee, J. B. et al. DNA primase acts as a molecular brake in DNA replication. Nature 439, 621–624 (2006)
Tanner, N. A. et al. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nature Struct. Mol. Biol. 15, 170–176 (2008)
Hamdan, S. M. et al. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol. Cell 27, 539–549 (2007)
van Oijen, A. M. et al. Single-molecule kinetics of lambda exonuclease reveal base dependence and dynamic disorder. Science 301, 1235–1238 (2003)
Lee, J., Chastain, P. D., Kusakabe, T., Griffith, J. D. & Richardson, C. C. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Mol. Cell 1, 1001–1010 (1998)
Frick, D. N. & Richardson, C. C. DNA primases. Annu. Rev. Biochem. 70, 39–80 (2001)
Qimron, U., Lee, S. J., Hamdan, S. M. & Richardson, C. C. Primer initiation and extension by T7 DNA primase. EMBO J. 25, 2199–2208 (2006)
Kusakabe, T. & Richardson, C. C. Gene 4 DNA primase of bacteriophage T7 mediates the annealing and extension of ribo-oligonucleotides at primase recognition sites. J. Biol. Chem. 272, 12446–12453 (1997)
Frick, D. N., Kumar, S. & Richardson, C. C. Interaction of ribonucleoside triphosphates with the gene 4 primase of bacteriophage T7. J. Biol. Chem. 274, 35899–35907 (1999)
Notarnicola, S. M., Mulcahy, H. L., Lee, J. & Richardson, C. C. The acidic carboxyl terminus of the bacteriophage T7 gene 4 helicase/primase interacts with T7 DNA polymerase. J. Biol. Chem. 272, 18425–18433 (1997)
Tabor, S., Huber, H. E. & Richardson, C. C. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J. Biol. Chem. 262, 16212–16223 (1987)
Hyland, E. M., Rezende, L. F. & Richardson, C. C. The DNA binding domain of the gene 2.5 single-stranded DNA-binding protein of bacteriophage T7. J. Biol. Chem. 278, 7247–7256 (2003)
Acknowledgements
We thank J.-B. Lee for technical advice and S. Moskowitz for illustrations. This work was supported by the National Institutes of Health (grants GM-077248 to A.M.v.O. and GM-54397 to C.C.R.) and the National Science Foundation (CAREER grant 0543784 to A.M.v.O.). J.J.L. acknowledges the Jane Coffin Childs Memorial Fund for a postdoctoral fellowship.
Author Contributions S.M.H. performed the single-molecule bead experiments; S.M.H. and J.J.L. performed the single-molecule fluorescence experiments; S.M.H. and M.T. performed the bulk-phase experiments; S.M.H., C.C.R. and A.M.v.O. designed the experiments, analysed the data and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Figures S1-S8 with Legends and Supplementary References. (PDF 1452 kb)
Rights and permissions
About this article
Cite this article
Hamdan, S., Loparo, J., Takahashi, M. et al. Dynamics of DNA replication loops reveal temporal control of lagging-strand synthesis. Nature 457, 336–339 (2009). https://doi.org/10.1038/nature07512
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07512
This article is cited by
-
Truncated TALE-FP as DNA Staining Dye in a High-salt Buffer
Scientific Reports (2019)
-
When proteins play tag: the dynamic nature of the replisome
Biophysical Reviews (2019)
-
Self-replication of DNA by its encoded proteins in liposome-based synthetic cells
Nature Communications (2018)
-
Helicase promotes replication re-initiation from an RNA transcript
Nature Communications (2018)
-
Freezing shortens the lifetime of DNA molecules under tension
Journal of Biological Physics (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.