Abstract
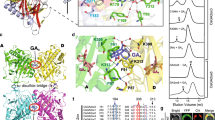
Gibberellins control a range of growth and developmental processes in higher plants and have been widely used in the agricultural industry. By binding to a nuclear receptor, GIBBERELLIN INSENSITIVE DWARF1 (GID1), gibberellins regulate gene expression by promoting degradation of the transcriptional regulator DELLA proteins, including GIBBERELLIN INSENSITIVE (GAI). The precise manner in which GID1 discriminates and becomes activated by bioactive gibberellins for specific binding to DELLA proteins remains unclear. Here we present the crystal structure of a ternary complex of Arabidopsis thaliana GID1A, a bioactive gibberellin and the amino-terminal DELLA domain of GAI. In this complex, GID1A occludes gibberellin in a deep binding pocket covered by its N-terminal helical switch region, which in turn interacts with the DELLA domain containing DELLA, VHYNP and LExLE motifs. Our results establish a structural model of a plant hormone receptor that is distinct from the mechanism of the hormone perception and effector recognition of the known auxin receptors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Yabuta, T. & Sumiki, Y. On the crystal of gibberellin, a substance to promote plant growth. J. Agric. Chem. Soc. Japan 14, 1526 (1938)
Radley, M. Occurrence of substances similar to gibberellic acid in higher plants. Nature 178, 1070–1071 (1956)
Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251 (2008)
MacMillan, J. Occurrence of gibberellins in vascular plants, fungi, and bacteria. J. Plant Growth Regul. 20, 387–442 (2002)
Ueguchi-Tanaka, M. et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698 (2005)
Nakajima, M. et al. Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46, 880–889 (2006)
Peng, J. et al. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205 (1997)
Silverstone, A. L., Ciampaglio, C. N. & Sun, T.-p. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10, 155–169 (1998)
McGinnis, K. M. et al. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15, 1120–1130 (2003)
Sasaki, A. et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299, 1896–1898 (2003)
Dill, A., Jung, H.-S. & Sun, T.-p. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl Acad. Sci. USA 98, 14162–14167 (2001)
Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M. & Matsuoka, M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14, 57–70 (2002)
Griffiths, J. et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis . Plant Cell 18, 3399–3414 (2006)
Willige, B. C. et al. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis . Plant Cell 19, 1209–1220 (2007)
Ueguchi-Tanaka, M. et al. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19, 2140–2155 (2007)
Pysh, L. D., Wysocka-Diller, J. W., Camilleri, C., Bouchez, D. & Benfey, P. N. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18, 111–119 (1999)
Silverstone, A. L. et al. Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis . Plant Cell 13, 1555–1565 (2001)
Dill, A., Thomas, S. G., Hu, J., Steber, C. M. & Sun, T.-p. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16, 1392–1405 (2004)
Fu, X. et al. The Arabidopsis mutant sleepy1gar2–1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16, 1406–1418 (2004)
Zentella, R. et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis . Plant Cell 19, 3037–3057 (2007)
Iuchi, S. et al. Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J. 50, 958–966 (2007)
Talon, M., Koornneef, M. & Zeevaart, J. A. D. Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc. Natl Acad. Sci. USA 87, 7983–7987 (1990)
Derkx, M. P. M., Vermeer, E. & Karssen, C. M. Gibberellins in seeds of Arabidopsis thaliana: biological activities, identification and effects of light and chilling on endogenous levels. Plant Growth Regul. 15, 223–234 (1994)
King, K. E., Moritz, T. & Harberd, N. P. Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159, 767–776 (2001)
Dill, A. & Sun, T.-p. Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana . Genetics 159, 777–785 (2001)
Lee, S. et al. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following inhibition. Genes Dev. 16, 646–658 (2002)
Cheng, H. et al. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131, 1055–1064 (2004)
Tyler, L. et al. DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis . Plant Physiol. 135, 1008–1019 (2004)
Ileperuma, N. R. et al. High-resolution crystal structure of plant carboxylesterase AeCXE1, from Actinidia eriantha, and its complex with a high-affinity inhibitor paraoxon. Biochemistry 46, 1851–1859 (2007)
Thomas, S. G., Phillips, A. L. & Hedden, P. Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl Acad. Sci. USA 96, 4698–4703 (1999)
Varbanova, M. et al. Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19, 32–45 (2007)
Crick, F. H. C. The packing of α-helices: simple coiled coils. Acta Crystallogr. 6, 689–697 (1953)
Hakoshima, T. in Nature Encyclopedia of the Human Genome Vol. 3, 679–683 (Nature Publishing Group, 2003)
McSteen, P. & Zhao, Y. Plant hormones and signaling: common themes and new developments. Dev. Cell 14, 467–473 (2008)
Ho, M. S., Ou, C., Chan, Y.-r., Chien, C.-T. & Pi, H. The utility F-box for protein destruction. Cell. Mol. Life Sci. 65, 1977–2000 (2008)
Dharmasiri, N., Dharmasiri, S. & Estelle, M. The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445 (2005)
Kepinski, S. & Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451 (2005)
Thines, B. et al. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448, 661–665 (2007)
Chini, A. et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671 (2007)
Tan, X. et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645 (2007)
Alberts, B. et al. in Molecular Biology of the Cell 5th edn, 395–396 (Garland Science, 2008)
LeMaster, D. M. & Richards, F. M. 1H–15N heteronuclear NMR studies of Escherichia coli thioredoxin in samples isotopically labeled by residue type. Biochemistry 24, 7263–7268 (1985)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Weeks, C. M. et al. Towards automated protein structure determination: BnP, the SnB-PHASES interface. Z. Kristallogr. 217, 686–693 (2002)
Terwilliger, T. SOLVE and RESOLVE: Automated structure solution, density modification and model building. J. Synchrotron Radiat. 11, 49–52 (2004)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Brünger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007)
Weiner, M. P. et al. Site-directed mutagenesis of double-strand DNA by the polymerase chain reaction. Gene 151, 119–123 (1994)
Acknowledgements
We thank S. Terawaki and T. Mori for technical advice and J. Tsukamoto for technical support in performing the matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy analysis, and the beamline staff of BL41XU of SPring-8 at Harima for help with the data collection. We also acknowledge A. Isogai for advice and encouragement. This work was supported by a research grant in the natural sciences from the Mitsubishi Foundation, Japan (to T.H.), and in part by a Grant-in-Aid for Scientific Research (A) and Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (to T.H.). This work was also supported in part by grants from the National Science Foundation of the United States (IBN-0348814 to T.-p.S.). K.M. is a recipient of a postdoctoral fellowship for Young Scientists from the Japan Society for the Promotion of Science and an international research fellowship from the Global COE Program in NAIST (Frontier Biosciences: strategies for survival and adaptation in a changing global environment) from MEXT of Japan.
Author Contributions K.M., T.-p.S. and T.H. conceived and designed the project. T.-p.S. provided all cDNA and constructs as well as expertise in the GA signalling mechanism. Y.H., K.M. and T.H. were responsible for construct design for protein preparation. K.M. was responsible for subcloning and execution of protein biochemistry, crystallization and data collection, helped by Y.H. and directed by T.H. Y.H. solved and refined the complex structures. K.M. performed binding studies with GID1A and DELLA variants, as well as circular dichroism measurements, directed by Y.H. Y.H, K.M. and T.H. interpreted data and T.H. wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1 and 2 and Supplementary Figures S1-S8 with Legends. (PDF 3182 kb)
Rights and permissions
About this article
Cite this article
Murase, K., Hirano, Y., Sun, Tp. et al. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463 (2008). https://doi.org/10.1038/nature07519
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature07519
This article is cited by
-
A tomato NAC transcription factor, SlNAP1, directly regulates gibberellin-dependent fruit ripening
Cellular & Molecular Biology Letters (2024)
-
The master growth regulator DELLA binding to histone H2A is essential for DELLA-mediated global transcription regulation
Nature Plants (2023)
-
Identification of a new gibberellin receptor agonist, diphegaractin, by a cell-free chemical screening system
Communications Biology (2023)
-
Analysis of the Physiological Roles and Mode of Actions of Phthalimides as GA Signal Regulator in Rice
Journal of Plant Growth Regulation (2023)
-
A rice seed-specific glycine-rich protein OsDOR1 interacts with GID1 to repress GA signaling and regulates seed dormancy
Plant Molecular Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.