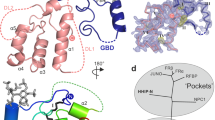
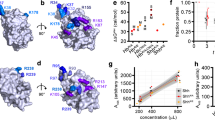
Abstract
Hedgehog (Hh) proteins specify tissue pattern in metazoan embryos by forming gradients that emanate from discrete sites of expression and elicit concentration-dependent cellular differentiation or proliferation responses1,2. Cellular responses to Hh and the movement of Hh through tissues are both precisely regulated, and abnormal Hh signalling has been implicated in human birth defects and cancer3,4,5,6,7. Hh signalling is mediated by its amino-terminal domain (HhN), which is dually lipidated and secreted as part of a multivalent lipoprotein particle8,9,10. Reception of the HhN signal is modulated by several cell-surface proteins on responding cells, including Patched (Ptc), Smoothened (Smo), Ihog (known as CDO or CDON in mammals) and the vertebrate-specific proteins Hip (also known as Hhip) and Gas1 (ref. 11). Drosophila Ihog and its vertebrate homologues CDO and BOC contain multiple immunoglobulin and fibronectin type III (FNIII) repeats, and the first FNIII repeat of Ihog binds Drosophila HhN in a heparin-dependent manner12,13. Surprisingly, pull-down experiments suggest that a mammalian Sonic hedgehog N-terminal domain (ShhN) binds a non-orthologous FNIII repeat of CDO12,14. Here we report biochemical, biophysical and X-ray structural studies of a complex between ShhN and the third FNIII repeat of CDO. We show that the ShhN–CDO interaction is completely unlike the HhN–Ihog interaction and requires calcium, which binds at a previously undetected site on ShhN. This site is conserved in nearly all Hh proteins and is a hotspot for mediating interactions between ShhN and CDO, Ptc, Hip and Gas1. Mutations in vertebrate Hh proteins causing holoprosencephaly and brachydactyly type A1 map to this calcium-binding site and disrupt interactions with these partners.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ingham, P. W. & McMahon, A. P. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15, 3059–3087 (2001)
Hooper, J. E. & Scott, M. P. Communicating with Hedgehogs. Nature Rev. Mol. Cell Biol. 6, 306–317 (2005)
Belloni, E. et al. Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nature Genet. 14, 353–356 (1996)
Roessler, E. et al. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nature Genet. 14, 357–360 (1996)
Gao, B. & He, L. Answering a century old riddle: brachydactyly type A1. Cell Res. 14, 179–187 (2004)
Hahn, H. et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell 85, 841–851 (1996)
Johnson, R. L. et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 272, 1668–1671 (1996)
Mann, R. K. & Beachy, P. A. Novel lipid modifications of secreted protein signals. Annu. Rev. Biochem. 73, 891–923 (2004)
Panakova, D., Sprong, H., Marois, E., Thiele, C. & Eaton, S. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature 435, 58–65 (2005)
Chen, M.-H., Li, Y.-J., Kawakami, T., Xu, S.-M. & Chuang, P.-T. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 18, 641–659 (2004)
Kang, J. S., Zhang, W. & Krauss, R. S. Hedgehog signaling: cooking with Gas1. Sci. STKE 403, pe50 (2007)
Yao, S., Lum, L. & Beachy, P. The Ihog cell-surface proteins bind Hedgehog and mediate pathway activation. Cell 125, 343–357 (2006)
McLellan, J. S. et al. Structure of a heparin-dependent complex of Hedgehog and Ihog. Proc. Natl Acad. Sci. USA 103, 17208–17213 (2006)
Tenzen, T. et al. The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Dev. Cell 10, 647–656 (2006)
Kang, J.-S. et al. CDO: an oncogene-, serum-, and anchorage-regulated member of the Ig/fibronectin type III repeat family. J. Cell Biol. 138, 203–213 (1997)
Conte, L. L., Chothia, C. & Janin, J. The atomic structure of protein–protein recognition sites. J. Mol. Biol. 285, 2177–2198 (1999)
Lawrence, M. C. & Colman, P. M. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234, 946–950 (1993)
Okada, A. et al. Boc is a receptor for sonic hedgehog in the guidance of commissural axons. Nature 444, 369–373 (2006)
Chimento, D. P., Mohanty, A. K., Kadner, R. J. & Wiener, M. C. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nature Struct. Mol. Biol. 10, 394–401 (2003)
Harding, M. M. The geometry of metal–ligand interactions relevant to proteins. Acta Crystallogr. D 55, 1432–1443 (1999)
Hall, T. M. T., Porter, J. A., Beachy, P. A. & Leahy, D. J. A potential catalytic site revealed by the 1.7-A crystal structure of the amino-terminal signalling domain of Sonic hedgehog. Nature 378, 212–216 (1995)
Kang, J.-S., Mulieri, P. J., Hu, Y., Taliana, L. & Krauss, R. S. BOC, an Ig superfamily member, associates with CDO to positively regulate myogenic differentiation. EMBO J. 21, 114–124 (2002)
Martinelli, D. C. & Fan, C.-M. Gas1 extends the range of Hedgehog action by facilitating its signaling. Genes Dev. 21, 1231–1243 (2007)
Heussler, H. S., Suri, M., Young, I. D. & Muenke, M. Extreme variability of expression of a Sonic Hedgehog mutation: attention difficulties and holoprosencephaly. Arch. Dis. Child. 86, 293–296 (2002)
Allen, B. L., Tenzen, T. & McMahon, A. P. The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 21, 1244–1257 (2007)
Chuang, P.-T. & McMahon, A. P. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature 397, 617–621 (1999)
Niedermaier, M. et al. An inversion involving the mouse Shh locus results in brachydactyly through dysregulation of Shh expression. J. Clin. Invest. 115, 900–909 (2005)
Kuriyan, J. & Eisenberg, D. The origin of protein interactions and allostery in colocalization. Nature 450, 983–990 (2007)
Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 (2000)
Geisbrecht, B. V., Bouyain, S. & Pop, M. An optimized system for expression and purification of secreted bacterial proteins. Protein Expres. Purif. 46, 23–32 (2006)
Ma, Y. et al. Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell 111, 63–75 (2002)
Otwinowski, Z. & Minor, W. in Methods in Enzymology (eds Carter, C. W. Jr & Sweet, R. M.) 307–326 (Academic, 1997)
The Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Brunger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Lebowitz, J., Lewis, M. S. & Schuck, P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 11, 2067–2079 (2002)
Garcia de la Torre, J., Huertas, M. L. & Carrasco, B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 78, 719–730 (2000)
Del Sal, G., Ruaro, M. E., Philipson, L. & Schneider, C. The growth arrest-specific gene, gas1, is involved in growth suppression. Cell 70, 595–607 (1992)
Acknowledgements
We thank R. Abramowitz and J. Schwanof of beamline X4C at the National Synchroton Light Source for assistance with X-ray data collection, C. W. Vander Kooi for suggesting ions may promote Shh–CDO interactions, W. Yang, J. Nathans, W. I. Weis, K. C. Garcia, P. A. Cole and L. M. Amzel for comments on the manuscript, and D. Chan and K. Cheah for communicating results before publication. We thank A. P. McMahon and C. M. Fan for the mouse Hip1 and mouse Gas1–Fc complementary DNAs, respectively. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases (R.G.). D.J.L. is supported by R01 HD055545 and P.A.B. is an HHMI investigator. J.S.M. is supported by a National Science Foundation Graduate Research Fellowship. X.Z. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-1915-06).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains supplementary Figures 1-18 with Legends and Supplementary Tables 1-2 (PDF 2698 kb)
Rights and permissions
About this article
Cite this article
McLellan, J., Zheng, X., Hauk, G. et al. The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla. Nature 455, 979–983 (2008). https://doi.org/10.1038/nature07358
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07358
This article is cited by
-
Suppression of apoptosis impairs phalangeal joint formation in the pathogenesis of brachydactyly type A1
Nature Communications (2024)
-
Coordination of canonical and noncanonical Hedgehog signalling pathways mediated by WDR11 during primordial germ cell development
Scientific Reports (2023)
-
Association of Sonic Hedgehog with the extracellular matrix requires its zinc-coordination center
BMC Molecular and Cell Biology (2021)
-
Deletion of 2 amino acids in IHH in a Japanese family with brachydactyly type A1
BMC Medical Genomics (2021)
-
ZSWIM8 is a myogenic protein that partly prevents C2C12 differentiation
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.