Abstract
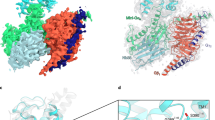
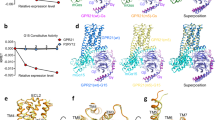
Epac proteins are activated by binding of the second messenger cAMP and then act as guanine nucleotide exchange factors for Rap proteins1,2. The Epac proteins are involved in the regulation of cell adhesion3 and insulin secretion4. Here we have determined the structure of Epac2 in complex with a cAMP analogue (Sp-cAMPS) and RAP1B by X-ray crystallography and single particle electron microscopy. The structure represents the cAMP activated state of the Epac2 protein with the RAP1B protein trapped in the course of the exchange reaction. Comparison with the inactive conformation reveals that cAMP binding causes conformational changes that allow the cyclic nucleotide binding domain to swing from a position blocking the Rap binding site towards a docking site at the Ras exchange motif domain.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Primary accessions
Protein Data Bank
Data deposits
Co-ordinates have been deposited with the Protein Data Bank under accession number 3cf6. The EM map of activated full length Epac2 bound to RAP1B has been deposited in the 3D EM database (http://www.ebi.ac.uk/msd/) under accession code EMD-1510.
References
de Rooij, J. et al. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature 396, 474–477 (1998)
Kawasaki, H. et al. A family of cAMP-binding proteins that directly activate Rap1. Science 282, 2275–2279 (1998)
Bos, J. L. Linking Rap to cell adhesion. Curr. Opin. Cell Biol. 17, 123–128 (2005)
Seino, S. & Shibasaki, T. PKA-dependent and PKA-independent pathways for cAMP-regulated exocytosis. Physiol. Rev. 85, 1303–1342 (2005)
Rehmann, H., Das, J., Knipscheer, P., Wittinghofer, A. & Bos, J. L. Structure of the cyclic-AMP-responsive exchange factor Epac2 in its auto-inhibited state. Nature 439, 625–628 (2006)
Rehmann, H., Schwede, F., Døskeland, S. O., Wittinghofer, A. & Bos, J. L. Ligand-mediated activation of the cAMP-responsive guanine nucleotide exchange factor Epac. J. Biol. Chem. 278, 38548–38556 (2003)
Canaves, J. M. & Taylor, S. S. Classification and phylogenetic analysis of the cAMP-dependent protein kinase regulatory subunit family. J. Mol. Evol. 54, 17–19 (2002)
Yagura, T. S. & Miller, J. P. Mapping adenosine cyclic 3′,5′-phosphate binding sites on type I and type II adenosine cyclic 3′,5′-phosphate dependent protein kinases using ribose ring and cyclic phosphate ring analogues of adenosine cyclic 3′,5′-phosphate. Biochemistry (Mosc.) 20, 879–887 (1981)
Su, Y. et al. Regulatory subunit of protein kinase A: Structure of deletion mutant with cAMP binding domains. Science 269, 807–813 (1995)
Diller, T. C., Madhusudan, N. H. & Taylor, S. S. Molecular basis for regulatory subunit diversity in cAMP-dependent protein kinase: Crystal structure of the type II beta regulatory subunit. Structure (Camb.) 9, 73–82 (2001)
Clayton, G. M., Silverman, W. R., Heginbotham, L. & Morais-Cabral, J. H. Structural basis of ligand activation in a cyclic nucleotide regulated potassium channel. Cell 119, 615–627 (2004)
Zagotta, W. N. et al. Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature 425, 200–205 (2003)
Rehmann, H., Wittinghofer, A. & Bos, J. L. Capturing cyclic nucleotides in action: Snapshots from crystallographic studies. Nature Rev. Mol. Cell Biol. 8, 63–73 (2007)
Margarit, S. M. et al. Structural evidence for feedback activation by RaS˙GTP of the Ras-specific nucleotide exchange factor SOS. Cell 112, 685–695 (2003)
Freedman, T. S. et al. A Ras-induced conformational switch in the Ras activator Son of sevenless. Proc. Natl Acad. Sci. USA 103, 16692–16697 (2006)
Boriack-Sjodin, P. A., Margarit, S. M., Bar-Sagi, D. & Kuriyan, J. The structural basis of the activation of Ras by Sos. Nature 394, 337–343 (1998)
Rehmann, H. Characterization of the activation of the Rap-specific exchange factor Epac by cyclic nucleotides. Methods Enzymol. 407, 159–173 (2006)
Herrmann, C., Horn, G., Spaargaren, M. & Wittinghofer, A. Differential interaction of the ras family GTP-binding proteins H-Ras, Rap1A, and R-Ras with the putative effector molecules Raf kinase and Ral-guanine nucleotide exchange factor. J. Biol. Chem. 271, 6794–6800 (1996)
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993)
Vagin, A. & Teplyakov, A. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D 56, 1622–1624 (2000)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Kraulis, P. J. Molscript — a program to produced both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991)
Schomburg, D. & Reichelt, J. Bragi — a comprehensive protein modeling program system. J. Mol. Graphics 6, 161–165 (1988)
Merritt, E. A. & Murphy, M. E. P. Rasert3D version 2.0 — A program for photorealistic molecular graphics. Acta Crystallogr. D 50, 869–873 (1994)
Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Heymann, J. B. & Belnap, D. M. Bsoft: Image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157, 3–18 (2007)
Scheres, S. H. et al. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nature Methods 4, 27–29 (2007)
Scheres, S. H. et al. Maximum-likelihood multi-reference refinement for electron microscopy images. J. Mol. Biol. 348, 139–149 (2005)
Sorzano, C. O. et al. XMIPP: A new generation of an open-source image processing package for electron microscopy. J. Struct. Biol. 148, 194–204 (2004)
Pettersen, E. F. et al. UCSF Chimera — a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Wriggers, W., Milligan, R. A. & McCammon, J. A. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 125, 185–195 (1999)
Garzon, J. I., Kovacs, J., Abagyan, R. & Chacon, P. ADP_EM: Fast exhaustive multi-resolution docking for high-throughput coverage. Bioinformatics 23, 427–433 (2007)
Frank, J. et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996)
Acknowledgements
We thank P. Gros for access to crystallization robots, M. Rensen-De Leeuw for technical assistance, A. Wittinghofer for reading the manuscript, the European Synchrotron Radiation Facility for providing synchrotron facilities, the scientists at ID23-1 for help with data collection, and the Galicia Supercomputer Centre (CESGA) and the Barcelona Supercomputing Centre for computing resources. H.R. is a recipient of the Hendrik Casimir-Karl Ziegler-Forschungspreis of the Nordrhein-Westfälischen Akademie der Wissenschaften and the Koninklijke Nederlandse Akademie van Wetenschappen. E.A.-P. and O.L. are supported by the Autonomous Region of Madrid, F.S. by the Bremer Innovationsagentur, O.L. by the Spanish Ministry of Education and Science (MEC) and the Red Temática de Investigación cooperativa en Cáncer (RTICC), and J.L.B. by the Chemical Sciences and the Netherlands Genomic Initiative of the Netherlands Organisation for Scientific Research (NWO).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information 1
This file contains Supplementary Table 1 describing the data collections and refinement statistics, a legend to Supplementary Movie describing the content of the movie in detail and Supplementary Figures S1 to S9 showing detail aspect of the EM and the crystal structures as well as a biochemical data. (PDF 5084 kb)
Supplementary Information 2
This file contains Supplementary Movie 1. This movie illustrates in cartoon style how cAMP induces the activation of Epac. The conformational changes within the CNB domain are shown left and the resulting structural consequences for the domain organisation of Epac right. A detailed description can be found in the Supplementary Information file. (MOV 2324 kb)
Rights and permissions
About this article
Cite this article
Rehmann, H., Arias-Palomo, E., Hadders, M. et al. Structure of Epac2 in complex with a cyclic AMP analogue and RAP1B. Nature 455, 124–127 (2008). https://doi.org/10.1038/nature07187
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07187
This article is cited by
-
Membranes prime the RapGEF EPAC1 to transduce cAMP signaling
Nature Communications (2023)
-
Sensitive genetically encoded sensors for population and subcellular imaging of cAMP in vivo
Nature Methods (2022)
-
Mutated RAS-associating proteins and ERK activation in relapse/refractory diffuse large B cell lymphoma
Scientific Reports (2022)
-
Modeling Epac1 interactions with the allosteric inhibitor AM-001 by co-solvent molecular dynamics
Journal of Computer-Aided Molecular Design (2020)
-
Insights into exchange factor directly activated by cAMP (EPAC) as potential target for cancer treatment
Molecular and Cellular Biochemistry (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.