Abstract
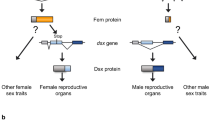
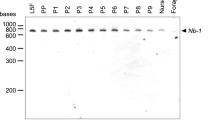
Sex determination in honeybees (Apis mellifera) is governed by heterozygosity at a single locus harbouring the complementary sex determiner (csd) gene1, in contrast to the well-studied sex chromosome system of Drosophila melanogaster2. Bees heterozygous at csd are females, whereas homozygotes and hemizygotes (haploid individuals) are males. Although at least 15 different csd alleles are known among natural bee populations3, the mechanisms linking allelic interactions to switching of the sexual development programme are still obscure. Here we report a new component of the sex-determining pathway in honeybees, encoded 12 kilobases upstream of csd. The gene feminizer (fem) is the ancestrally conserved progenitor gene from which csd arose and encodes an SR-type protein, harbouring an Arg/Ser-rich domain. Fem shares the same arrangement of Arg/Ser- and proline-rich-domain with the Drosophila principal sex-determining gene transformer (tra), but lacks conserved motifs except for a 30-amino-acid motif that Fem shares only with Tra of another fly, Ceratitis capitata4. Like tra, the fem transcript is alternatively spliced. The male-specific splice variant contains a premature stop codon and yields no functional product, whereas the female-specific splice variant encodes the functional protein. We show that RNA interference (RNAi)-induced knockdowns of the female-specific fem splice variant result in male bees, indicating that the fem product is required for entire female development. Furthermore, RNAi-induced knockdowns of female allelic csd transcripts result in the male-specific fem splice variant, suggesting that the fem gene implements the switch of developmental pathways controlled by heterozygosity at csd. Comparative analysis of fem and csd coding sequences from five bee species indicates a recent origin of csd in the honeybee lineage from the fem progenitor and provides evidence for positive selection at csd accompanied by purifying selection at fem. The fem locus in bees uncovers gene duplication and positive selection as evolutionary mechanisms underlying the origin of a novel sex determination pathway.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Primary accessions
GenBank/EMBL/DDBJ
Data deposits
The sequences generated in this study are available from GenBank under the accession numbers EU101387 (GB11211), EU101388 (femF), EU101389 (femM), EU101390 (csd), EU101391 (GB30480), EU101392 (GB13727), EU139305 (M. compressipes fem), EU288185 (B. terrestris fem), and EU100885–EU100941 (Apis fem and csd sequences from populations and different species). SDL assembly and sequence annotation data are available in the Third Party Annotation Section of the DDBJ/EMBL/GenBank databases under the accession number TPA: BK006346.
References
Beye, M., Hasselmann, M., Fondrk, M. K., Page, R. E. & Omholt, S. W. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell 114, 419–429 (2003)
Cline, T. W. & Meyer, B. J. Vive la différence: males vs females in flies vs worms. Annu. Rev. Genet. 30, 637–702 (1996)
Hasselmann, M. & Beye, M. Signatures of selection among sex-determining alleles of the honey bee. Proc. Natl Acad. Sci. USA 101, 4888–4893 (2004)
Pane, A., Salvemini, M., Bovi, P. D., Polito, C. & Saccone, G. The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development 129, 3715–3725 (2002)
Hasselmann, M. & Beye, M. Pronounced differences of recombination activity at the sex determination locus (SDL) of the honey bee, a locus under strong balancing selection. Genetics 174, 1469–1480 (2006)
Honeybee Genome Sequencing Consortium Insights into social insects from the genome of the honeybee Apis mellifera . Nature 443, 931–949 (2006)
Blencowe, B. J., Bowman, J. A., McCracken, S. & Rosonina, E. SR-related proteins and the processing of messenger RNA precursors. Biochem. Cell Biol. 77, 277–291 (1999)
Kulathinal, R. J., Skwarek, L., Morton, R. A. & Singh, R. S. Rapid evolution of the sex-determining gene, transformer: structural diversity and rate heterogeneity among sibling species of Drosophila . Mol. Biol. Evol. 20, 441–452 (2003)
Butler, B., Pirrotta, V., Irminger-Finger, I. & Nothiger, R. The sex-determining gene tra of Drosophila: molecular cloning and transformation studies. EMBO J. 5, 3607–3613 (1986)
Boggs, R. T., Gregor, P., Idriss, S., Belote, J. M. & McKeown, M. Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell 50, 739–747 (1987)
Bell, L. R., Maine, E. M., Schedl, P. & Cline, T. W. Sex-lethal, a Drosophila sex determination switch gene, exhibits sex-specific RNA splicing and sequence similarity to RNA binding proteins. Cell 55, 1037–1046 (1988)
Keyes, L. N., Cline, T. W. & Schedl, P. The primary sex determination signal of Drosophila acts at the level of transcription. Cell 68, 933–943 (1992)
Hoshijima, K., Inoue, K., Higuchi, I., Sakamoto, H. & Shimura, Y. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila . Science 252, 833–836 (1991)
Grimaldi, D. & Engel, M. S. Evolution of the Insects 454–467 (Cambridge Univ. Press, UK, 2005)
Kerr, W. E. Sex determination in bees. XXI. Number of XO-heteroalleles in a natural population of Melipona compressipes fasciculata (Apidae). Insectes Soc. 34, 274–279 (1987)
Cook, J. M. Sex determination in the Hymenoptera: a review of models and evidence. Heredity 71, 421–435 (1993)
Burkhard, P., Stetefeld, J. & Strelkov, S. V. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11, 82–88 (2001)
Beye, M. The dice of fate: the csd gene and how its allelic composition regulates sexual development in the honey bee, Apis mellifera . Bioessays 26, 1131–1139 (2004)
Wilkins, A. S. Moving up the hierarchy: A hypothesis on the evolution of a genetic sex determination pathway. Bioessays 17, 71–77 (1995)
Nöthiger, R. & Steinemann-Zwicky, M. A single principle for sex determination in insects. Cold Spring Harb. Symp. Quant. Biol. 50, 615–621 (1985)
Bull, J. J. Evolution of Sex Determining Mechanisms (Benjamin/Cummings Publishing Company, Menlo Park, California, 1983)
Pomiankowski, A., Nothiger, R. & Wilkins, A. The evolution of the Drosophila sex-determination pathway. Genetics 166, 1761–1773 (2004)
Hill, W. G. & Robertson, A. The effect of linkage on limits to artificial selection. Genet. Res. 8, 269–294 (1966)
Otto, S. P. & Lenormand, T. Resolving the paradox of sex and recombination. Nature Rev. Genet. 3, 252–261 (2002)
Lewis, D. The evolution of sex in flowering plants. Biol. Rev. 17, 46–67 (1942)
Charlesworth, B. & Charlesworth, D. A model for the evolution of dioecy and gynodioecy. Am. Nat. 112, 975–997 (1978)
Charlesworth, D., Charlesworth, B. & Marais, G. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95, 118–128 (2005)
Graves, J. A. Sex chromosome specialization and degeneration in mammals. Cell 124, 901–914 (2006)
Nei, M. & Kumar, S. Molecular Evolution and Phylogenetics 216–221 (Oxford Univ. Press, New York, 2000)
Engel, M. S. Monophyly and extensive extinction of advanced eusocial bees: insights from an unexpected Eocene diversity. Proc. Natl Acad. Sci. USA 98, 1661–1664 (2001)
Acknowledgements
We thank W. Martin, A. Wilkins, T. Eltz and J. Baines for comments on the manuscript; E.-M. Theilenberg, M. Mueller-Borg and C. Schulte for technical support; D. Titera for providing bee crosses; J. Pflugfelder, N. Koeniger, G. Koeniger, J. Bozic and S. Tingek for collecting honeybee samples; K. Lunau for providing bumble bee samples; and M. Griese for bee-keeping support. This work was supported by grants from the Deutsche Forschungsgemeinschaft DFG.
Author Contributions M.H. performed the evolutionary nucleotide analysis, isolated gene sequences from different species and supervised some experiments; T.G. performed the gene studies; M.S. assembled SDL sequences and identified genes; C.G.N.-S. characterized M. compressipes sequences; M.O. analysed domain structures; and M.B. performed the experimental design, supervised the research project and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-2, Supplementary Tables 1-2 and Supplementary Notes. (PDF 359 kb)
Rights and permissions
About this article
Cite this article
Hasselmann, M., Gempe, T., Schiøtt, M. et al. Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature 454, 519–522 (2008). https://doi.org/10.1038/nature07052
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07052
This article is cited by
-
The function and evolution of a genetic switch controlling sexually dimorphic eye differentiation in honeybees
Nature Communications (2023)
-
Abundant small RNAs in the reproductive tissues and eggs of the honey bee, Apis mellifera
BMC Genomics (2022)
-
Transcriptome analysis reveals differentially expressed genes between the ovary and testis of the honey bee Apis mellifera
Apidologie (2022)
-
The honey bee genome-- what has it been good for?
Apidologie (2021)
-
Characterization of sex-specific variants of doublesex and feminizer genes in stingless bee species
Apidologie (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.