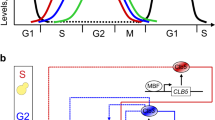
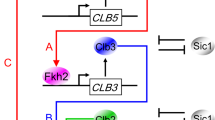
Abstract
In budding yeast, Saccharomyces cerevisiae, the Start checkpoint integrates multiple internal and external signals into an all-or-none decision to enter the cell cycle. Here we show that Start behaves like a switch due to systems-level feedback in the regulatory network. In contrast to current models proposing a linear cascade of Start activation, transcriptional positive feedback of the G1 cyclins Cln1 and Cln2 induces the near-simultaneous expression of the ∼200-gene G1/S regulon. Nuclear Cln2 drives coherent regulon expression, whereas cytoplasmic Cln2 drives efficient budding. Cells with the CLN1 and CLN2 genes deleted frequently arrest as unbudded cells, incurring a large fluctuation-induced fitness penalty due to both the lack of cytoplasmic Cln2 and insufficient G1/S regulon expression. Thus, positive-feedback-amplified expression of Cln1 and Cln2 simultaneously drives robust budding and rapid, coherent regulon expression. A similar G1/S regulatory network in mammalian cells, comprised of non-orthologous genes, suggests either conservation of regulatory architecture or convergent evolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Simchen, G., Pinon, R. & Salts, Y. Sporulation in Saccharomyces cerevisiae: premeiotic DNA synthesis, readiness and commitment. Exp. Cell Res. 75, 207–218 (1972)
Nachman, I., Regev, A. & Ramanathan, S. Dissecting timing variability in yeast meiosis. Cell 131, 544–556 (2007)
Shenhar, G. & Kassir, Y. A positive regulator of mitosis, Sok2, functions as a negative regulator of meiosis in Saccharomyces cerevisiae . Mol. Cell. Biol. 21, 1603–1612 (2001)
Ferrell, J. E. & Machleder, E. M. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science 280, 895–898 (1998)
Xiong, W. & Ferrell, J. E. A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature 426, 460–465 (2003)
Sha, W. et al. Hysteresis drives cell-cycle transitions in Xenopus laevis egg extracts. Proc. Natl Acad. Sci. USA 100, 975–980 (2003)
Pomerening, J. R., Sontag, E. D. & Ferrell, J. E. Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nature Cell Biol. 5, 346–351 (2003)
Hartwell, L. H., Culotti, J., Pringle, J. R. & Reid, B. J. Genetic control of the cell division cycle in yeast. Science 183, 46–51 (1974)
Johnston, G. C., Pringle, J. R. & Hartwell, L. H. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae . Exp. Cell Res. 105, 79–98 (1977)
Lord, P. G. & Wheals, A. E. Variability in individual cell cycles of Saccharomyces cerevisiae . J. Cell Sci. 50, 361–376 (1981)
Di Talia, S., Skotheim, J. M., Bean, J. M., Siggia, E. D. & Cross, F. R. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature 448, 947–951 (2007)
Jorgensen, P. & Tyers, M. How cells coordinate growth and division. Curr. Biol. 14, R1014–R1027 (2004)
Tyers, M., Tokiwa, G. & Futcher, B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 12, 1955–1968 (1993)
Dirick, L., Bohm, T. & Nasmyth, K. Roles and regulation of Cln–Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae . EMBO J. 14, 4803–4813 (1995)
Stuart, D. & Wittenberg, C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 9, 2780–2794 (1995)
Spellman, P. T. et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9, 3273–3297 (1998)
Kato, M., Hata, N., Banerjee, N., Futcher, B. & Zhang, M. Q. Identifying combinatorial regulation of transcription factors and binding motifs. Genome Biol. 5, R56 (2004)
de Bruin, R. A., McDonald, W. H., Kalashnikova, T. I., Yates, J. & Wittenberg, C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117, 887–898 (2004)
Costanzo, M. et al. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117, 899–913 (2004)
Amon, A., Tyers, M., Futcher, B. & Nasmyth, K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell 74, 993–1007 (1993)
de Bruin, R. A. et al. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol. Cell 23, 483–496 (2006)
Cross, F. R. & Tinkelenberg, A. H. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell 65, 875–883 (1991)
Dirick, L. & Nasmyth, K. Positive feedback in the activation of G1 cyclins in yeast. Nature 351, 754–757 (1991)
Bean, J. M., Siggia, E. D. & Cross, F. R. Coherence and timing of cell cycle Start examined at single-cell resolution. Mol. Cell 21, 3–14 (2006)
Mateus, C. & Avery, S. V. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast 16, 1313–1323 (2000)
Samoilov, M. S., Price, G. & Arkin, A. P. From fluctuations to phenotypes: the physiology of noise. Sci. STKE 2006, re17 (2006)
Iyer, V. R. et al. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409, 533–538 (2001)
Harbison, C. T. et al. Transcriptional regulatory code of a eukaryotic genome. Nature 431, 99–104 (2004)
Simon, I. et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106, 697–708 (2001)
Koch, C., Schleiffer, A., Ammerer, G. & Nasmyth, K. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at Start, whereas Clb/Cdc28 kinases displace it from the promoter in G2 . Genes Dev. 10, 129–141 (1996)
Moffat, J. & Andrews, B. Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast. Nature Cell Biol. 6, 59–66 (2004)
Zachariae, W., Schwab, M., Nasmyth, K. & Seufert, W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 282, 1721–1724 (1998)
Wijnen, H., Landman, A. & Futcher, B. The G1 cyclin Cln3 promotes cell cycle entry via the transcription factor Swi6. Mol. Cell. Biol. 22, 4402–4418 (2002)
Edgington, N. P. & Futcher, B. Relationship between the function and the location of G1 cyclins in S. cerevisiae . J. Cell Sci. 114, 4599–4611 (2001)
Miller, M. E. & Cross, F. R. Distinct subcellular localization patterns contribute to functional specificity of the Cln2 and Cln3 cyclins of Saccharomyces cerevisiae . Mol. Cell. Biol. 20, 542–555 (2000)
Koch, C., Moll, T., Neuberg, M., Ahorn, H. & Nasmyth, K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science 261, 1551–1557 (1993)
Bean, J. M., Siggia, E. D. & Cross, F. R. High functional overlap between MBF and SBF in the G1/S transcriptional program in Saccharomyces cerevisiae . Genetics 171, 49–61 (2005)
McCusker, D. et al. Cdk1 coordinates cell-surface growth with the cell cycle. Nature Cell Biol. 9, 506–515 (2007)
Polymenis, M. & Schmidt, E. V. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 11, 2522–2531 (1997)
Wang, H., Gari, E., Verges, E., Gallego, C. & Aldea, M. Recruitment of Cdc28 by Whi3 restricts nuclear accumulation of the G1 cyclin–Cdk complex to late G1. EMBO J. 23, 180–190 (2004)
Schneider, B. L., Yang, Q. H. & Futcher, A. B. Linkage of replication to Start by the Cdk inhibitor Sic1. Science 272, 560–562 (1996)
Lanker, S., Valdivieso, M. H. & Wittenberg, C. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science 271, 1597–1601 (1996)
Shaner, N. C. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnol. 22, 1567–1572 (2004)
Acknowledgements
This work was supported by the National Institute of Health (J.M.S., E.D.S. and F.R.C.), the Burroughs Wellcome Fund (J.M.S.) and the National Science Foundation (E.D.S.). We thank N. Buchler, G. Charvin, B. Drapkin and J. E. Ferrell for conversations; J. Widom and C. Wittenberg for comments on the manuscript; J. M. Bean, B. Timney and J. Robbins for help with strain/plasmid construction; M. Schwab for the plasmid pWS358; B. Futcher for the CLN2-NES and CLN2-NLS plasmids; E. Bi for the pKT355 mCherry tagging plasmid; and M. Tyers for WHI5 phosphorylation site mutant strains and plasmids.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary information
The file contains Supplementary Methods, Supplementary Table and Supplementary Figures S1-S14 with Legends. (PDF 1575 kb)
Rights and permissions
About this article
Cite this article
Skotheim, J., Di Talia, S., Siggia, E. et al. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature 454, 291–296 (2008). https://doi.org/10.1038/nature07118
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07118
This article is cited by
-
The CDK Pho85 inhibits Whi7 Start repressor to promote cell cycle entry in budding yeast
EMBO Reports (2024)
-
Non-coding RNAs and angiogenesis in cardiovascular diseases: a comprehensive review
Molecular and Cellular Biochemistry (2024)
-
Multidimensional characterization of inducible promoters and a highly light-sensitive LOV-transcription factor
Nature Communications (2023)
-
Imperatorin induces autophagy and G0/G1 phase arrest via PTEN-PI3K-AKT-mTOR/p21 signaling pathway in human osteosarcoma cells in vitro and in vivo
Cancer Cell International (2021)
-
Cyclin/Forkhead-mediated coordination of cyclin waves: an autonomous oscillator rationalizing the quantitative model of Cdk control for budding yeast
npj Systems Biology and Applications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.