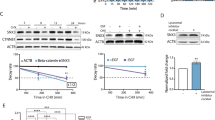
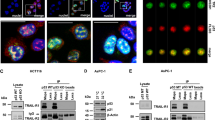
Abstract
NUMB is a cell fate determinant, which, by asymmetrically partitioning at mitosis, controls cell fate choices by antagonising the activity of the plasma membrane receptor of the NOTCH family1. NUMB is also an endocytic protein2, and the NOTCH–NUMB counteraction has been linked to this function3,4. There might be, however, additional functions of NUMB, as witnessed by its proposed role as a tumour suppressor in breast cancer5. Here we describe a previously unknown function for human NUMB as a regulator of tumour protein p53 (also known as TP53). NUMB enters in a tricomplex with p53 and the E3 ubiquitin ligase HDM2 (also known as MDM2), thereby preventing ubiquitination and degradation of p53. This results in increased p53 protein levels and activity, and in regulation of p53-dependent phenotypes. In breast cancers there is frequent loss of NUMB expression5. We show that, in primary breast tumour cells, this event causes decreased p53 levels and increased chemoresistance. In breast cancers, loss of NUMB expression causes increased activity of the receptor NOTCH5. Thus, in these cancers, a single event—loss of NUMB expression—determines activation of an oncogene (NOTCH) and attenuation of the p53 tumour suppressor pathway. Biologically, this results in an aggressive tumour phenotype, as witnessed by findings that NUMB-defective breast tumours display poor prognosis. Our results uncover a previously unknown tumour suppressor circuitry.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Roegiers, F. & Jan, Y. N. Asymmetric cell division. Curr. Opin. Cell Biol. 16, 195–205 (2004)
Santolini, E. et al. Numb is an endocytic protein. J. Cell Biol. 151, 1345–1352 (2000)
Berdnik, D., Torok, T., Gonzalez-Gaitan, M. & Knoblich, J. A. The endocytic protein α-adaptin is required for numb-mediated asymmetric cell division in Drosophila . Dev. Cell 3, 221–231 (2002)
Hutterer, A. & Knoblich, J. A. Numb and α-adaptin regulate Sanpodo endocytosis to specify cell fate in Drosophila external sensory organs. EMBO Rep. 6, 836–842 (2005)
Pece, S. et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 167, 215–221 (2004)
Vousden, K. H. & Prives, C. p53 and prognosis: new insights and further complexity. Cell 120, 7–10 (2005)
Hollstein, M., Sidransky, D., Vogelstein, B. & Harris, C. C. p53 mutations in human cancers. Science 253, 49–53 (1991)
Momand, J., Zambetti, G. P., Olson, D. C., George, D. & Levine, A. J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69, 1237–1245 (1992)
Haupt, Y., Maya, R., Kazaz, A. & Oren, M. Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 (1997)
Kubbutat, M. H., Jones, S. N. & Vousden, K. H. Regulation of p53 stability by Mdm2. Nature 387, 299–303 (1997)
Zhang, Y., Xiong, Y. & Yarbrough, W. G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92, 725–734 (1998)
Oliner, J. D., Kinzler, K. W., Meltzer, P. S., George, D. L. & Vogelstein, B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358, 80–83 (1992)
Sherr, C. J. Tumor surveillance via the ARF–p53 pathway. Genes Dev. 12, 2984–2991 (1998)
McCann, A. H. et al. Amplification of the MDM2 gene in human breast cancer and its association with MDM2 and p53 protein status. Br. J. Cancer 71, 981–985 (1995)
Pharoah, P. D., Day, N. E. & Caldas, C. Somatic mutations in the p53 gene and prognosis in breast cancer: a meta-analysis. Br. J. Cancer 80, 1968–1973 (1999)
Vestey, S. B. et al. p14ARFexpression in invasive breast cancers and ductal carcinoma in situ—relationships to p53 and Hdm2. Breast Cancer Res. 6, R571–R585 (2004)
Sharpless, N. E. & DePinho, R. A. The INK4A/ARF locus and its two gene products. Curr. Opin. Genet. Dev. 9, 22–30 (1999)
Silva, J. et al. Analysis of genetic and epigenetic processes that influence p14ARF expression in breast cancer. Oncogene 20, 4586–4590 (2001)
Silva, J. et al. Concomitant expression of p16INK4a and p14ARF in primary breast cancer and analysis of inactivation mechanisms. J. Pathol. 199, 289–297 (2003)
Juven-Gershon, T. et al. The Mdm2 oncoprotein interacts with the cell fate regulator Numb. Mol. Cell. Biol. 18, 3974–3982 (1998)
Vassilev, L. T. et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 (2004)
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 (1998)
Paull, T. T. et al. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10, 886–895 (2000)
Reis, T. & Edgar, B. A. Negative regulation of dE2F1 by cyclin-dependent kinases controls cell cycle timing. Cell 117, 253–264 (2004)
Lowe, S. W., Ruley, H. E., Jacks, T. & Housman, D. E. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell 74, 957–967 (1993)
Fish, K. N., Schmid, S. L. & Damke, H. Evidence that dynamin-2 functions as a signal-transducing GTPase. J. Cell Biol. 150, 145–154 (2000)
Enari, M., Ohmori, K., Kitabayashi, I. & Taya, Y. Requirement of clathrin heavy chain for p53-mediated transcription. Genes Dev. 20, 1087–1099 (2006)
Sharpless, N. E. & DePinho, R. A. Telomeres, stem cells, senescence, and cancer. J. Clin. Invest. 113, 160–168 (2004)
Sherley, J. L., Stadler, P. B. & Johnson, D. R. Expression of the wild-type p53 antioncogene induces guanine nucleotide-dependent stem cell division kinetics. Proc. Natl Acad. Sci. USA 92, 136–140 (1995)
Rambhatla, L., Ram-Mohan, S., Cheng, J. J. & Sherley, J. L. Immortal DNA strand cosegregation requires p53/IMPDH-dependent asymmetric self-renewal associated with adult stem cells. Cancer Res. 65, 3155–3161 (2005)
Rubinson, D. A. et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nature Genet. 33, 401–406 (2003)
Woelk, T. et al. Molecular mechanisms of coupled monoubiquitination. Nature Cell Biol. 8, 1246–1254 (2006)
Veronesi, U. et al. A randomized comparison of sentinel-node biopsy with routine axillary dissection in breast cancer. N. Engl. J. Med. 349, 546–553 (2003)
Acknowledgements
We thank K. Helin for the p53 and HDM2 reagents; L. Van Parijis for the pLL3.7 lentiviral vector; R. Bernard for the p53 shRNA pSUPER vector; G. Matera for technical assistance; P. Maisonneuve and G. Goisis for statistical analysis; the Imaging Service at IEO; and the Real Time PCR Service at IFOM. This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro and MIUR to S.P. and P.P.D.F., from the European Community (VI Framework), The Ferrari Foundation, the Monzino Foundation and the CARIPLO Foundation to P.P.D.F., and from the G. Vollaro Foundation to S.P.
Author Contributions I.N.C., D.T., F.S.-N. and S.P. performed experimental work. P.N., V.G. and G.V. performed the clinical part of the work (patient selection, histology and data analysis of the patient’s case collection). S.P. and P.P.D.F. planned and supervised the project, performed data analysis and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
This file contains Supplementary Discussion and additional references; Supplementary Table S1 and Supplementary Figures S1-S6 with Legends. (PDF 3132 kb)
Rights and permissions
About this article
Cite this article
Colaluca, I., Tosoni, D., Nuciforo, P. et al. NUMB controls p53 tumour suppressor activity. Nature 451, 76–80 (2008). https://doi.org/10.1038/nature06412
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature06412
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.