Abstract
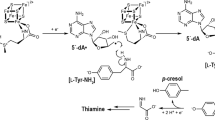
Eubacterial leucyl/phenylalanyl-tRNA protein transferase (LF-transferase) catalyses peptide-bond formation by using Leu-tRNALeu (or Phe-tRNAPhe) and an amino-terminal Arg (or Lys) of a protein, as donor and acceptor substrates, respectively. However, the catalytic mechanism of peptide-bond formation by LF-transferase remained obscure. Here we determine the structures of complexes of LF-transferase and phenylalanyl adenosine, with and without a short peptide bearing an N-terminal Arg. Combining the two separate structures into one structure as well as mutation studies reveal the mechanism for peptide-bond formation by LF-transferase. The electron relay from Asp 186 to Gln 188 helps Gln 188 to attract a proton from the α-amino group of the N-terminal Arg of the acceptor peptide. This generates the attacking nucleophile for the carbonyl carbon of the aminoacyl bond of the aminoacyl-tRNA, thus facilitating peptide-bond formation. The protein-based mechanism for peptide-bond formation by LF-transferase is similar to the reverse reaction of the acylation step observed in the peptide hydrolysis reaction by serine proteases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Varshavsky, A. The N-end rule. Cell 69, 725–735 (1992)
Varshavsky, A. The N-end rule: functions, mysteries, uses. Proc. Natl Acad. Sci. USA 93, 12142–12149 (1996)
Mogk, A., Schmidt, R. & Bukau, R. The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol. 17, 165–172 (2007)
Rao, H., Uhlmann, F., Nasmyth, K. & Varshavsky, A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature 410, 955–959 (2001)
Varshavsky, A. The N-end rule and regulation of apoptosis. Nature Cell Biol. 5, 373–376 (2003)
Hu, R. G. et al. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 437, 981–986 (2005)
Varshavsky, A. The ubiquitin system. Trends Biochem. Sci. 22, 383–387 (1997)
Nandi, D., Tahiliani, P., Kumar, A. & Chandu, D. The ubiquitin–proteasome system. J. Biosci. 31, 137–155 (2006)
Dougan, D. A., Mogk, A., Zeth, K., Turgay, K. & Bukaku, B. AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett. 529, 6–10 (2002)
Erbse, A. et al. ClpS is an essential component of the N-end rule pathway in Escherichia coli . Nature 439, 753–756 (2006)
Tobias, J. W., Shrader, T. E., Rocap, G. & Varshavsky, A. The N-end rule in bacteria. Science 254, 1374–1377 (1991)
Hegde, S. S. & Shrader, T. E. FemABX family members are novel nonribosomal peptidyltransferases and important pathogen-specific drug targets. J. Biol. Chem. 276, 6998–7003 (2001)
Benson, T. E. et al. X-ray crystal structure of Staphylococcus aureus FemA. Structure 10, 1107–1115 (2002)
Biarrotte-Sorin, S. et al. Crystal structures of Weissella viridescens FemX and its complex with UDP-MurNAc-pentapeptide: insights into FemABX family substrates recognition. Structure 12, 257–267 (2004)
Vetting, M. W. et al. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433, 212–226 (2005)
Kaji, H., Novelli, G. D. & Kaji, A. A soluble amino acid-incorporating system from rat liver. Biochim. Biophys. Acta 76, 474–477 (1963)
Soffer, R. L. Peptide acceptors in the leucine, phenylalanine transfer reaction. J. Biol. Chem. 248, 8424–8428 (1973)
Ichetovkin, I. E., Abramochkin, G. & Shrader, T. E. Substrate recognition by the leucyl/phenylalanyl-tRNA-protein transferase. Conservation within the enzyme family and localization to the trypsin-resistant domain. J. Biol. Chem. 272, 33009–33014 (1997)
Suto, K. et al. Crystal structures of leucyl/phenylalanyl-tRNA-protein transferase and its complex with an aminoacyl-tRNA analog. EMBO J. 25, 5942–5950 (2006)
Eriani, G, Delarue, M., Poch, O., Gangloff, J. & Moras, D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 347, 203–206 (1990)
Abramochkin, G. & Shrader, T. E. Aminoacyl-tRNA recognition by the leucyl/phenylalanyl-tRNA-protein transferase. J. Biol. Chem. 271, 22901–22907 (1996)
Blow, D. M., Briktoft, J. J. & Hartley, B. S. Role of a buried acid group in the mechanism of action of chymotrypsin. Nature 221, 337–340 (1969)
Steitz, T. A. & Shulman, R. G. Crystallographic and NMR studies of the serine proteases. Annu. Rev. Biophys. Bioeng. 11, 419–444 (1982)
Nissen, P., Hansen, J., Ban, N., Moore, P. B. & Steitz, T. A. The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 (2000)
Beringer, M. & Rodnina, M. V. The ribosomal peptidyl transferase. Mol. Cell 26, 311–321 (2007)
Rodnina, M. V., Beringer, M. & Wintermeyer, W. How ribosomes make peptide bonds. Trends Biochem. Sci. 32, 20–26 (2007)
Schmeing, T. M., Huang, K. S., Kitchen, D. E., Strobel, S. A. & Steitz, T. A. Structural insights into the roles of water and the 2' hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol. Cell 20, 437–448 (2005)
Trobro, S. & Åqvist, J. Analysis of predictions for the catalytic mechanism of ribosomal peptidyl transfer. Biochemistry 45, 7049–7056 (2006)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Cowtan, K. An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM. Newslett. Protein Crystallogr. 31, 34–38 (1994)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Gottikh, B. P., Krayevsky, A. A., Tarussova, N. B., Purygin, P. P. & Tsilevich, T. L. The general synthetic route to amino acid esters of nucleotides and nucleoside-5′-triphosphates and some properties of these compounds. Tetrahedron 26, 4419–4433 (1970)
Chládek, S., Ringer, D. & Žemlička, J. L-Phenylalanine esters of open-chain analog of adenosine as substrates for ribosomal peptidyl transferase. Biochemistry 12, 5135–5138 (1973)
Roy, H., Ling, J., Irnov, M. & Ibba, M. Post-transfer editing in vitro and in vivo by the β subunit of phenylalanyl-tRNA synthetase. EMBO J. 23, 4639–4648 (2004)
Acknowledgements
We thank S. Fukai, T. Numata, T. Suzuki and H. Hori for valuable and critical comments, and suggestions for this manuscript. We thank the beam-line staffs at BL-5A, BL-17A and AR-NW 12A of KEK (Tsukuba, Japan) for technical help during data collection, and A. Hamada for technical assistance. This work was supported in part by grants from JSPS, MEXT and the Kurata Memorial Hitachi Science and Technology Foundation (K.T.).
Author Contributions K.T. purified and crystallized the proteins, and K.W., Y.T. and K.T. collected the data and determined the structures. K.S. assisted with the structural analysis, K.W. and K.T. carried out biochemical and mass analyses, Y.S. assisted with the mass analysis, and N.O. and T.W. synthesized analogues. K.W., Y.T. and K.T. wrote the paper. All authors discussed the results and commented on the manuscript.
Coordinates and structure factors have been deposited in the Protein Data Bank, under the accession codes 2Z3K, 2Z3L, 2Z3M, 2Z3N, 2Z3O and 2Z3P.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Discussion and additional references, Supplementary Table S1 and Supplementary Figures S1-S9 with Legends. (PDF 7940 kb)
Rights and permissions
About this article
Cite this article
Watanabe, K., Toh, Y., Suto, K. et al. Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase. Nature 449, 867–871 (2007). https://doi.org/10.1038/nature06167
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06167
This article is cited by
-
Mechanism of aminoacyl-tRNA acetylation by an aminoacyl-tRNA acetyltransferase AtaT from enterohemorrhagic E. coli
Nature Communications (2020)
-
Leveraging nature’s biomolecular designs in next-generation protein sequencing reagent development
Applied Microbiology and Biotechnology (2020)
-
A consistent force field parameter set for zwitterionic amino acid residues
Journal of Molecular Modeling (2014)
-
The N-end rule pathway: emerging functions and molecular principles of substrate recognition
Nature Reviews Molecular Cell Biology (2011)
-
Protein Microarrays: Novel Developments and Applications
Pharmaceutical Research (2011)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.