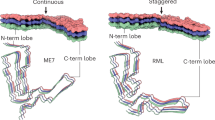
Abstract
Among the many surprises to arise from studies of prion biology, perhaps the most unexpected is the strain phenomenon whereby a single protein can misfold into structurally distinct, infectious states that cause distinguishable phenotypes1,2,3. Similarly, proteins can adopt a spectrum of conformations in non-infectious diseases of protein folding; some are toxic and others are well tolerated4. However, our understanding of the structural differences underlying prion strains and how these differences alter their physiological impact remains limited. Here we use a combination of solution NMR, amide hydrogen/deuterium (H/D) exchange and mutagenesis to study the structural differences between two strain conformations, termed Sc4 and Sc37 (ref. 5), of the yeast Sup35 prion. We find that these two strains have an overlapping amyloid core spanning most of the Gln/Asn-rich first 40 amino acids that is highly protected from H/D exchange and very sensitive to mutation. These features indicate that the cores are composed of tightly packed β-sheets possibly resembling ‘steric zipper’ structures revealed by X-ray crystallography of Sup35-derived peptides6,7. The stable structure is greatly expanded in the Sc37 conformation to encompass the first 70 amino acids, revealing why this strain shows increased fibre stability and decreased ability to undergo chaperone-mediated replication8. Our findings establish that prion strains involve large-scale conformational differences and provide a structural basis for understanding a broad range of functional studies, including how conformational changes alter the physiological impact of prion strains.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Cohen, F. E. & Prusiner, S. B. Pathologic conformations of prion proteins. Annu. Rev. Biochem. 67, 793–819 (1998)
Derkatch, I. L., Chernoff, Y. O., Kushnirov, V. V., Inge-Vechtomov, S. G. & Liebman, S. W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144, 1375–1386 (1996)
Tuite, M. F. & Cox, B. S. The [PSI+] prion of yeast: a problem of inheritance. Methods 39, 9–22 (2006)
Chiti, F. & Dobson, C. M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006)
Tanaka, M., Chien, P., Naber, N., Cooke, R. & Weissman, J. S. Conformational variations in an infectious protein determine prion strain differences. Nature 428, 323–328 (2004)
Nelson, R. et al. Structure of the cross-β spine of amyloid-like fibrils. Nature 435, 773–778 (2005)
Sawaya, M. R. et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 447, 453–457 (2007)
Tanaka, M., Collins, S. R., Toyama, B. H. & Weissman, J. S. The physical basis of how prion conformations determine strain phenotypes. Nature 442, 585–589 (2006)
Glover, J. R. et al. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89, 811–819 (1997)
Sparrer, H. E., Santoso, A., Szoka, F. C. & Weissman, J. S. Evidence for the prion hypothesis: induction of the yeast [PSI+] factor by in vitro-converted Sup35 protein. Science 289, 595–599 (2000)
King, C. Y. & Diaz-Avalos, R. Protein-only transmission of three yeast prion strains. Nature 428, 319–323 (2004)
Krishnan, R. & Lindquist, S. L. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435, 765–772 (2005)
Shewmaker, F., Wickner, R. B. & Tycko, R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc. Natl Acad. Sci. USA 103, 19754–19759 (2006)
Liu, J. J., Sondheimer, N. & Lindquist, S. L. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion. Proc. Natl Acad. Sci. USA [PSI+]. 99 (suppl. 4). 16446–16453 (2002)
Flaux, J., Bertelsen, E. B., Horwich, A. L. & Wuthrich, K. NMR analysis of a 900K GroEL–GroES complex. Nature 418, 207–211 (2002)
Hoshino, M. et al. Mapping the core of the β2-microglobulin amyloid fibril by H/D exchange. Nature Struct. Biol. 9, 332–336 (2002)
Yamaguchi, K. et al. Core and heterogeneity of β2-microglobulin amyloid fibrils as revealed by H/D exchange. J. Mol. Biol. 338, 559–571 (2004)
Ritter, C. et al. Correlation of structural elements and infectivity of the HET-s prion. Nature 435, 844–848 (2005)
Luhrs, T. et al. 3D structure of Alzheimer’s amyloid-β(1–42) fibrils. Proc. Natl Acad. Sci. USA 102, 17342–17347 (2005)
Carulla, N. et al. Molecular recycling within amyloid fibrils. Nature 436, 554–558 (2005)
Chien, P., DePace, A. H., Collins, S. R. & Weissman, J. S. Generation of prion transmission barriers by mutational control of amyloid conformations. Nature 424, 948–951 (2003)
DePace, A. H. & Weissman, J. S. Origins and kinetic consequences of diversity in Sup35 yeast prion fibers. Nature Struct. Biol. 9, 389–396 (2002)
Ross, E. D., Edskes, H. K., Terry, M. J. & Wickner, R. B. Primary sequence independence for prion formation. Proc. Natl Acad. Sci. USA 102, 12825–12830 (2005)
Tessier, P. M. & Lindquist, S. Prion recognition elements govern nucleation, strain specificity and species barriers. Nature 447, 556–561 (2007)
King, C. Y. Supporting the structural basis of prion strains: induction and identification of [PSI] variants. J. Mol. Biol. 307, 1247–1260 (2001)
DePace, A. H., Santoso, A., Hillner, P. & Weissman, J. S. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 93, 1241–1252 (1998)
Osherovich, L. Z., Cox, B. S., Tuite, M. F. & Weissman, J. S. Dissection and design of yeast prions. PLoS Biol. 2, E86 (2004)
Crist, C. G., Nakayashiki, T., Kurahashi, H. & Nakamura, Y. [PHI+], a novel Sup35-prion variant propagated with non-Gln/Asn oligopeptide repeats in the absence of the chaperone protein Hsp104. Genes Cells 8, 603–618 (2003)
Parham, S. N., Resende, C. G. & Tuite, M. F. Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions. EMBO J. 20, 2111–2119 (2001)
Shkundina, I. S., Kushnirov, V. V., Tuite, M. F. & Ter-Avanesyan, M. D. The role of the N-terminal oligopeptide repeats of the yeast Sup35 prion protein in propagation and transmission of prion variants. Genetics 172, 827–835 (2006)
Santoso, A., Chien, P., Osherovich, L. Z. & Weissman, J. S. Molecular basis of a yeast prion species barrier. Cell 100, 277–288 (2000)
Muchmore, D. C., McIntosh, L. P., Russell, C. B., Anderson, D. E. & Dahlquist, F. W. Expression and nitrogen-15 labeling of proteins for proton and nitrogen-15 nuclear magnetic resonance. Methods Enzymol. 177, 44–73 (1989)
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995)
Goddard, T. D. & Kneller, D. G. SPARKY 3.112, University of California, San Francisco. (2006)
Sattler, M., Schleucher, J. & Griesinger, C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc. 34, 93–158 (1999)
Sun, Z. Y., Frueh, D. P., Selenko, P., Hoch, J. C. & Wagner, G. Fast assignment of 15N-HSQC peaks using high-resolution 3D HNcocaNH experiments with non-uniform sampling. J. Biomol. NMR 33, 43–50 (2005)
Kraulis, P. J. ANSIG: A program for the assignment of protein 1H 2D NMR spectra by interactive graphics. J. Magn. Reson. 84, 627–633 (1989)
Acknowledgements
We thank M. Tanaka, C. Ritter, M. Hoshino, W. Bermel and R. Riek for experimental advice; and D. Breslow, D. Cameron, S. Collins, V. Denic, K. Filaski, C. Foo, C. Gross, J. Hollien, N. Ingolia, E. Quan, E. Rodriguez and K. Tipton and other members of the Weissman laboratory for helpful discussion and critical reading of the manuscript. This research was funded by the NIH and the Howard Hughes Medical Institute.
Backbone assignments of SupNM have been deposited in the Biological Magnetic Resonance Data Bank, accession number 15379.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures S1-S5 with Legends (PDF 1143 kb)
Rights and permissions
About this article
Cite this article
Toyama, B., Kelly, M., Gross, J. et al. The structural basis of yeast prion strain variants. Nature 449, 233–237 (2007). https://doi.org/10.1038/nature06108
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06108
This article is cited by
-
Amyloid conformation-dependent disaggregation in a reconstituted yeast prion system
Nature Chemical Biology (2022)
-
Atomic-level differences between brain parenchymal- and cerebrovascular-seeded Aβ fibrils
Scientific Reports (2021)
-
Protein assembly systems in natural and synthetic biology
BMC Biology (2020)
-
Amino acid homorepeats in proteins
Nature Reviews Chemistry (2020)
-
Short disordered protein segment regulates cross-species transmission of a yeast prion
Nature Chemical Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.