Abstract
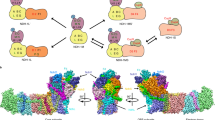
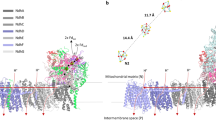
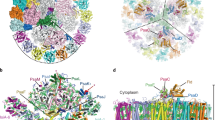
Oxygen-evolving photosynthetic organisms regulate carbon metabolism through a light-dependent redox signalling pathway1. Electrons are shuttled from photosystem I by means of ferredoxin (Fdx) to ferredoxin–thioredoxin reductase (FTR), which catalyses the two-electron-reduction of chloroplast thioredoxins (Trxs). These modify target enzyme activities by reduction, regulating carbon flow2. FTR is unique in its use of a [4Fe–4S] cluster and a proximal disulphide bridge in the conversion of a light signal into a thiol signal2. We determined the structures of FTR in both its one- and its two-electron-reduced intermediate states and of four complexes in the pathway, including the ternary Fdx–FTR–Trx complex. Here we show that, in the first complex (Fdx–FTR) of the pathway, the Fdx [2Fe–2S] cluster is positioned suitably for electron transfer to the FTR [4Fe–4S] centre. After the transfer of one electron, an intermediate is formed in which one sulphur atom of the FTR active site is free to attack a disulphide bridge in Trx and the other sulphur atom forms a fifth ligand for an iron atom in the FTR [4Fe–4S] centre—a unique structure in biology. Fdx then delivers a second electron that cleaves the FTR–Trx heterodisulphide bond, which occurs in the Fdx–FTR–Trx complex. In this structure, the redox centres of the three proteins are aligned to maximize the efficiency of electron transfer from the Fdx [2Fe–2S] cluster to the active-site disulphide of Trxs. These results provide a structural framework for understanding the mechanism of disulphide reduction by an iron–sulphur enzyme3 and describe previously unknown interaction networks for both Fdx and Trx (refs 4–6).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Buchanan, B. B. & Balmer, Y. Redox regulation: a broadening horizon. Annu. Rev. Plant Biol. 56, 187–220 (2005)
Schürmann, P. Redox signaling in the chloroplast: the ferredoxin/thioredoxin system. Antioxid. Redox Signal. 5, 69–78 (2003)
Johnson, D. C., Dean, D. R., Smith, A. D. & Johnson, M. K. Structure, function, and formation of biological iron–sulfur clusters. Annu. Rev. Biochem. 74, 247–281 (2005)
Kurisu, G. et al. Structure of the electron transfer complex between ferredoxin and ferredoxin-NADP+ reductase. Nature Struct. Biol. 8, 117–121 (2001)
Morales, R. et al. A redox-dependent interaction between two electron-transfer partners involved in photosynthesis. EMBO Rep. 1, 271–276 (2000)
Lennon, B. W., Williams, C. H. & Ludwig, M. L. Twists in catalysis: alternating conformations of Escherichia coli thioredoxin reductase. Science 289, 1190–1194 (2000)
Dai, S. et al. How does light regulate chloroplast enzymes? Structure–function studies of the ferredoxin/thioredoxin system. Q. Rev. Biophys. 33, 67–108 (2000)
van den Heuvel, R. H. et al. The active conformation of glutamate synthase and its binding to ferredoxin. J. Mol. Biol. 330, 113–128 (2003)
Xu, X. et al. Ferredoxin/ferredoxin–thioredoxin reductase complex: complete NMR mapping of the interaction site on ferredoxin by gallium substitution. FEBS Lett. 580, 6714–6720 (2006)
Onuchic, J. N., Beratan, D. N., Winkler, J. R. & Gray, H. B. Pathway analysis of protein electron-transfer reactions. Annu. Rev. Biophys. Biomol. Struct. 21, 349–377 (1992)
Dai, S., Schwendtmayer, C., Schürmann, P., Ramaswamy, S. & Eklund, H. Redox signaling in chloroplasts: cleavage of disulfides by an iron–sulfur cluster. Science 287, 655–658 (2000)
Knaff, D. B. in Advances in Photosynthesis (eds Ort, D. R. & Yocum, C. F.) 333–361 (Kluwer Academic, Dordrecht, 1996)
Jameson, G. N. et al. Spectroscopic evidence for site specific chemistry at a unique iron site of the [4Fe–4S] cluster in ferredoxin:thioredoxin reductase. J. Am. Chem. Soc. 125, 1146–1147 (2003)
Staples, C. R. et al. Role of the [Fe4S4] cluster in mediating disulfide reduction in spinach ferredoxin:thioredoxin reductase. Biochemistry 37, 4612–4620 (1998)
Walters, E. M. et al. Spectroscopic characterization of site-specific [Fe4S4] cluster chemistry in ferredoxin:thioredoxin reductase: implications for the catalytic mechanism. J. Am. Chem. Soc. 127, 9612–9624 (2005)
Berkovitch, F., Nicolet, Y., Wan, J. T., Jarrett, J. T. & Drennan, C. L. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science 303, 76–79 (2004)
Hanzelmann, P. & Schindelin, H. Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc. Natl Acad. Sci. USA 101, 12870–12875 (2004)
Lauble, H., Kennedy, M. C., Beinert, H. & Stout, C. D. Crystal structures of aconitase with trans-aconitate and nitrocitrate bound. J. Mol. Biol. 237, 437–451 (1994)
Layer, G., Moser, J., Heinz, D. W., Jahn, D. & Schubert, W. D. Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of Radical SAM enzymes. EMBO J. 22, 6214–6224 (2003)
Lepore, B. W., Ruzicka, F. J., Frey, P. A. & Ringe, D. The X-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale. Proc. Natl Acad. Sci. USA 102, 13819–13824 (2005)
Staples, C. R. et al. The function and properties of the iron–sulfur center in spinach ferredoxin:thioredoxin reductase: a new biological role for iron–sulfur clusters. Biochemistry 35, 11425–11434 (1996)
Hirasawa, M. et al. Oxidation–reduction properties of chloroplast thioredoxins, ferredoxin:thioredoxin reductase, and thioredoxin f-regulated enzymes. Biochemistry 38, 5200–5205 (1999)
Giastas, P. et al. The structure of the 2[4Fe–4S] ferredoxin from Pseudomonas aeruginosa at 1.32-Å resolution: comparison with other high-resolution structures of ferredoxins and contributing structural features to reduction potential values. J. Biol. Inorg. Chem. 11, 445–458 (2006)
Buchanan, B. B., Schürmann, P., Decottignies, P. & Lozano, R. M. Thioredoxin: a multifunctional regulatory protein with a bright future in technology and medicine. Arch. Biochem. Biophys. 314, 257–260 (1994)
Glauser, D. A., Bourquin, F., Manieri, W. & Schürmann, P. Characterization of ferredoxin:thioredoxin reductase modified by site-directed mutagenesis. J. Biol. Chem. 279, 16662–16669 (2004)
Walters, E. M. & Johnson, M. K. Ferredoxin:thioredoxin reductase: disulfide reduction catalyzed via novel site-specific [4Fe–4S] cluster chemistry. Photosynth. Res. 79, 249–264 (2004)
Chen, K. et al. Crystal structures of ferredoxin variants exhibiting large changes in [Fe–S] reduction potential. Nature Struct. Biol. 9, 188–192 (2002)
Duin, E. C., Madadi-Kahkesh, S., Hedderich, R., Clay, M. D. & Johnson, M. K. Heterodisulfide reductase from Methanothermobacter marburgensis contains an active-site [4Fe–4S] cluster that is directly involved in mediating heterodisulfide reduction. FEBS Lett. 512, 263–268 (2002)
Chen, D., Walsby, C., Hoffman, B. M. & Frey, P. A. Coordination and mechanism of reversible cleavage of S-adenosylmethionine by the [4Fe–4S] center in lysine 2,3-aminomutase. J. Am. Chem. Soc. 125, 11788–11789 (2003)
Schürmann, P. & Gardet-Salvi, L. Chemical modification of the active site of ferredoxin–thioredoxin reductase. Chimia 47, 245–246 (1993)
Manieri, W. et al. N-terminal truncation of the variable subunit stabilizes spinach ferredoxin:thioredoxin reductase. FEBS Lett. 549, 167–170 (2003)
Balmer, Y. & Schürmann, P. Heterodimer formation between thioredoxin f and fructose 1,6-bisphosphatase from spinach chloroplasts. FEBS Lett. 492, 58–61 (2001)
Schürmann, P. Ferredoxin:thioredoxin system. Methods Enzymol. 252, 274–283 (1995)
Wedel, N., Clausmeyer, S., Herrmann, R. G., Gardet-Salvi, L. & Schürmann, P. Nucleotide sequence of cDNAs encoding the entire precursor polypeptide for thioredoxin m from spinach chloroplasts. Plant Mol. Biol. 18, 527–533 (1992)
Stura, E. A. in Crystallization of Nucleic Acids and Proteins. A Practical Approach (eds Ducruix, A. & Giegé, R.) 99–126 (Oxford Univ. Press, New York, 1999)
Schürmann, P., Stritt-Etter, A. L. & Li, J. Reduction of ferredoxin:thioredoxin reductase by artificial electron donors. Photosynth. Res. 46, 309–312 (1995)
Balmer, Y. et al. Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc. Natl Acad. Sci. USA 100, 370–375 (2003)
Brunger, A. T. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 (1992)
Brunger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Kleywegt, G. J., Henrick, K., Dodson, E. J. & van Aalten, D. M. Pound-wise but penny-foolish: how well do micromolecules fare in macromolecular refinement? Structure 11, 1051–1059 (2003)
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993)
Nicholls, A., Sharp, K. A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991)
DeLano, W. The PyMOL Molecular Graphics System. 〈http://www.pymol.org〉 (2002)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Navaza, J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 (1994)
Lelong, C., Setif, P., Bottin, H., Andre, F. & Neumann, J. M. 1H and 15N NMR sequential assignment, secondary structure, and tertiary fold of [2Fe–2S] ferredoxin from Synechocystis sp. PCC 6803. Biochemistry 34, 14462–14473 (1995)
Powell, H. R. The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta Crystallogr. D 55, 1690–1695 (1999)
Collaborative Computational Project No 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 (1997)
Acknowledgements
We thank J. Kappler, P. Marrack and J. Bolin for support and encouragement; the Zuckerman/Canyon Ranch and A. Lapporte for support of the X-ray and computing facilities; the Howard Hughes Medical Institute beamlines at Advanced Light Source (ALS), the Structural Biology Centre at Advanced Photon Source (APS), and the European Synchrotron Radiation Facility (ESRF) for synchrotron data. H.E. was supported by the Swedish Council for Forestry and Agricultural Research and the Swedish Natural Science Research Council, and P.S. was supported by the Schweizerischer Nationalfonds.
Author Contributions S.D., R.F., D.A.G., F.B., W.M. and P.S. performed the experiments. S.D., R.F., P.S. and H.E. designed and prepared the manuscript.
The atomic coordinates and structure factors of Fdx–FTR, NEM-FTR, two-electron-reduced FTR, FTR–Trx-f(C49S), Fdx–FTR–Trx-f(C49S) and FTR–Trx-m(C40S) have been deposited in the RCSB Protein Data Bank under accession codes 2PVG, 2PUO, 2PVD, 2PU9, 2PVO and 2PUK, respectively.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-9 and Supplementary Tables 1-2 with Legends and additional references. (PDF 4233 kb)
Rights and permissions
About this article
Cite this article
Dai, S., Friemann, R., Glauser, D. et al. Structural snapshots along the reaction pathway of ferredoxin–thioredoxin reductase. Nature 448, 92–96 (2007). https://doi.org/10.1038/nature05937
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature05937
This article is cited by
-
Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase
Nature Communications (2020)
-
X-ray structure of an asymmetrical trimeric ferredoxin–photosystem I complex
Nature Plants (2018)
-
Intra- and inter-protein couplings of backbone motions underlie protein thiol-disulfide exchange cascade
Scientific Reports (2018)
-
Roles and maturation of iron–sulfur proteins in plastids
JBIC Journal of Biological Inorganic Chemistry (2018)
-
Ferredoxin:thioredoxin reductase (FTR) links the regulation of oxygenic photosynthesis to deeply rooted bacteria
Planta (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.