Abstract
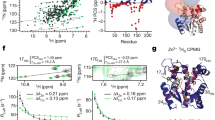
Protein folding and binding are analogous processes, in which the protein ‘searches’ for favourable intramolecular or intermolecular interactions on a funnelled energy landscape1,2. Many eukaryotic proteins are disordered under physiological conditions, and fold into ordered structures only on binding to their cellular targets3,4,5,6. The mechanism by which folding is coupled to binding is poorly understood, but it has been hypothesized on theoretical grounds that the binding kinetics may be enhanced by a ‘fly-casting’ effect, where the disordered protein binds weakly and non-specifically to its target and folds as it approaches the cognate binding site7. Here we show, using NMR titrations and 15N relaxation dispersion, that the phosphorylated kinase inducible activation domain (pKID) of the transcription factor CREB forms an ensemble of transient encounter complexes on binding to the KIX domain of the CREB binding protein. The encounter complexes are stabilized primarily by non-specific hydrophobic contacts, and evolve by way of an intermediate to the fully bound state without dissociation from KIX. The carboxy-terminal helix of pKID is only partially folded in the intermediate, and becomes stabilized by intermolecular interactions formed in the final bound state. Future applications of our method will provide new understanding of the molecular mechanisms by which intrinsically disordered proteins perform their diverse biological functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Tsai, C. J., Kumar, S., Ma, B. Y. & Nussinov, R. Folding funnels, binding funnels, and protein function. Protein Sci. 8, 1181–1190 (1999)
Levy, Y., Cho, S. S., Onuchic, J. N. & Wolynes, P. G. A survey of flexible protein binding mechanisms and their transition states using native topology based energy landscapes. J. Mol. Biol. 346, 1121–1145 (2005)
Wright, P. E. & Dyson, H. J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 293, 321–331 (1999)
Dyson, H. J. & Wright, P. E. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 12, 54–60 (2002)
Dyson, H. J. & Wright, P. E. Intrinsically unstructured proteins and their functions. Nature Rev. Mol. Cell Biol. 6, 197–208 (2005)
Uversky, V. N. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 11, 739–756 (2002)
Shoemaker, B. A., Portman, J. J. & Wolynes, P. G. Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc. Natl Acad. Sci. USA 97, 8868–8873 (2000)
Chrivia, J. C. et al. Phosphorylated CREB binds specifically to nuclear protein CBP. Nature 365, 855–859 (1993)
Kwok, R. P. S. et al. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370, 223–226 (1994)
Parker, D. et al. Phosphorylation of CREB at Ser133 induces complex formation with CPB via a direct mechanism. Mol. Cell. Biol. 16, 694–703 (1996)
Radhakrishnan, I. et al. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:coactivator interactions. Cell 91, 741–752 (1997)
Shih, H. M. et al. A positive genetic selection for disrupting protein-protein interactions: Identification of CREB mutations that prevent association with the coactivator CBP. Proc. Natl Acad. Sci. USA 93, 13896–13901 (1996)
Shaywitz, A. J., Dove, S. L., Kornhauser, J. M., Hochschild, A. & Greenberg, M. E. Magnitude of the CREB-dependent transcriptional response is determined by the strength of the interaction between the kinase-inducible domain of CREB and the KIX domain of CREB-binding protein. Mol. Cell. Biol. 20, 9409–9422 (2000)
Parker, D. et al. Analysis of an activator:coactivator complex reveals an essential role for secondary structure in transcriptional activation. Mol. Cell 2, 353–359 (1998)
Loria, J. P., Rance, M. & Palmer, A. G. A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 121, 2331–2332 (1999)
Mulder, F. A., Mittermaier, A., Hon, B., Dahlquist, F. W. & Kay, L. E. Studying excited states of proteins by NMR spectroscopy. Nature Struct. Biol. 8, 932–935 (2001)
Goto, N. K., Zor, T., Martinez-Yamout, M., Dyson, H. J. & Wright, P. E. Cooperativity in transcription factor binding to the coactivator CREB-binding protein (CBP). The mixed lineage leukemia protein (MLL) activation domain binds to an allosteric site on the KIX domain. J. Biol. Chem. 277, 43168–43174 (2002)
De Guzman, R. N., Goto, N. K., Dyson, H. J. & Wright, P. E. Structural basis for cooperative transcription factor binding to the CBP coactivator. J. Mol. Biol. 355, 1005–1013 (2006)
Gabdoulline, R. R. & Wade, R. C. Biomolecular diffusional association. Curr. Opin. Struct. Biol. 12, 204–213 (2002)
Berg, O. G. & von Hippel, P. H. Diffusion-controlled macromolecular interactions. Annu. Rev. Biophys. Biophys. Chem. 14, 131–160 (1985)
Tang, C., Iwahara, J. & Clore, G. M. Visualization of transient encounter complexes in protein-protein association. Nature 444, 383–386 (2006)
Volkov, A. N., Worrall, J. A. R., Holtzmann, E. & Ubbink, M. Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR. Proc. Natl Acad. Sci. USA 103, 18945–18950 (2006)
Zor, T., De Guzman, R. N., Dyson, H. J. & Wright, P. E. Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb. J. Mol. Biol. 337, 521–534 (2004)
Johnson, C. S. & Moreland, C. G. The calculation of NMR spectra for many-site exchange problems. J. Chem. Ed. 50, 477–483 (1973)
Shoup, D. & Szabo, A. Role of diffusion in ligand binding to macromolecules and cell-bound receptors. Biophys. J. 40, 33–39 (1982)
Selzer, T. & Schreiber, G. New insights into the mechanism of protein-protein association. Proteins Struct. Funct. Genet. 45, 190–198 (2001)
Tollinger, M., Skrynnikov, N. R., Mulder, F. A., Forman-Kay, J. D. & Kay, L. E. Slow dynamics in folded and unfolded states of an SH3 domain. J. Am. Chem. Soc. 123, 11341–11352 (2001)
McElheny, D., Schnell, J. R., Lansing, J. C., Dyson, H. J. & Wright, P. E. Defining the role of active-site loop fluctuations in dihydrofolate reductase catalysis. Proc. Natl Acad. Sci. USA 102, 5032–5037 (2005)
Koradi, R., Billeter, M. & Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55 (1996)
Huth, J. R. et al. Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. 6, 2359–2364 (1997)
Grey, M. J., Wang, C. & Palmer, A. G. Disulfide bond isomerization in basic pancreatic trypsin inhibitor: Multisite chemical exchange quantified by CPMG relaxation dispersion and chemical shift modeling. J. Am. Chem. Soc. 125, 14324–14335 (2003)
Carver, J. P. & Richards, R. E. General 2-site solution for chemical exchange produced dependence of T2 upon Carr-Purcell pulse separation. J. Magn. Reson. 6, 89–105 (1972)
Allerhand, A. & Gutowsky, H. S. Spin-echo studies of chemical exchange. II. Closed formulas for two sites. J. Chem. Phys. 42, 1587–1599 (1965)
Acknowledgements
We thank M. Martinez-Yamout for assistance with NMR titrations. This work was supported by the National Institutes of Health and the Skaggs Institute for Chemical Biology.
Author Contributions K.S. and P.E.W. designed the experiments, and K.S., P.E.W. and H.J.D. analysed the data and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures S1-S7 with Legends. (PDF 1178 kb)
Rights and permissions
About this article
Cite this article
Sugase, K., Dyson, H. & Wright, P. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 447, 1021–1025 (2007). https://doi.org/10.1038/nature05858
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05858
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.