Abstract
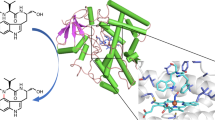
Enzyme-catalysed oxidations are some of the most common transformations in primary and secondary metabolism. The vancomycin biosynthetic enzyme DpgC belongs to a small class of oxygenation enzymes that are not dependent on an accessory cofactor or metal ion1. The detailed mechanism of cofactor-independent oxygenases has not been established. Here we report the first structure of an enzyme of this oxygenase class in complex with a bound substrate mimic. The use of a designed, synthetic substrate analogue allows unique insights into the chemistry of oxygen activation. The structure confirms the absence of cofactors, and electron density consistent with molecular oxygen is present adjacent to the site of oxidation on the substrate. Molecular oxygen is bound in a small hydrophobic pocket and the substrate provides the reducing power to activate oxygen for downstream chemical steps. Our results resolve the unique and complex chemistry of DpgC, a key enzyme in the biosynthetic pathway of an important class of antibiotics2. Furthermore, mechanistic parallels exist between DpgC and cofactor-dependent flavoenzymes3, providing information regarding the general mechanism of enzymatic oxygen activation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Tseng, C. C., Vaillancourt, F. H., Bruner, S. D. & Walsh, C. T. DpgC is a metal- and cofactor-free 3,5-dihydroxyphenylacetyl-CoA 1,2-dioxygenase in the vancomycin biosynthetic pathway. Chem. Biol. 11, 1195–1203 (2004)
Walsh, C. T. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science 303, 1805–1810 (2004)
Eswaramoorthy, S., Bonanno, J. B., Burley, S. K. & Swaminathan, S. Mechanism of action of a flavin-containing monooxygenase. Proc. Natl Acad. Sci. USA 103, 9832–9837 (2006)
Silverman, R. B. (ed.) The Organic Chemistry of Enzyme-Catalyzed Reactions. Revised edn. (Academic Press, San Diego, 2002)
Cane, D. E., Walsh, C. T. & Khosla, C. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science 282, 63–68 (1998)
Hubbard, B. K. & Walsh, C. T. Vancomycin assembly: nature's way. Angew. Chem. Int. Edn Engl. 42, 730–765 (2003)
Chen, H., Tseng, C. C., Hubbard, B. K. & Walsh, C. T. Glycopeptide antibiotic biosynthesis: enzymatic assembly of the dedicated amino acid monomer (S)-3,5-dihydroxyphenylglycine. Proc. Natl Acad. Sci. USA 98, 14901–14906 (2001)
Agnihotri, G. & Liu, H. W. Enoyl-CoA hydratase. reaction, mechanism, and inhibition. Bioorg. Med. Chem. 11, 9–20 (2003)
Holden, H. M., Benning, M. M., Haller, T. & Gerlt, J. A. The crotonase superfamily: divergently related enzymes that catalyze different reactions involving acyl coenzyme A thioesters. Acc. Chem. Res. 34, 145–157 (2001)
Beaman, T. W. et al. Acyl group specificity at the active site of tetrahydridipicolinate N-succinyltransferase. Protein Sci. 11, 974–979 (2002)
Xiang, H., Luo, L., Taylor, K. L. & Dunaway-Mariano, D. Interchange of catalytic activity within the 2-enoyl-coenzyme A hydratase/isomerase superfamily based on a common active site template. Biochemistry 38, 7638–7652 (1999)
Berglund, G. I. et al. The catalytic pathway of horseradish peroxidase at high resolution. Nature 417, 463–468 (2002)
Karlsson, A. et al. Crystal structure of naphthalene dioxygenase: side-on binding of dioxygen to iron. Science 299, 1039–1042 (2003)
Wentworth, P. et al. Antibody catalysis of the oxidation of water. Science 293, 1806–1811 (2001)
Adam, W., Alzerreca, A., Liu, J. C. & Yany, F. Cyclic peroxide 52. alpha-peroxylactones via dehydrative cyclization of alpha-hydroperoxy acids. J. Am. Chem. Soc. 99, 5768–5773 (1977)
Koo, J. A., Schmidt, S. P. & Schuster, G. B. Bioluminescence of the firefly: key steps in the formation of the electronically excited state for model systems. Proc. Natl Acad. Sci. USA 75, 30–33 (1978)
Fetzner, S. Oxygenases without requirement for cofactors or metal ions. Appl. Microbiol. Biotechnol. 60, 243–257 (2002)
Sciara, G. et al. The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis. EMBO J. 22, 205–215 (2003)
Colloc'h, N. et al. Crystal structure of the protein drug urate oxidase-inhibitor complex at 2.05 A resolution. Nature Struct. Biol. 4, 947–952 (1997)
Beinker, P. et al. Crystal structures of SnoaL2 and AclR: two putative hydroxylases in the biosynthesis of aromatic polyketide antibiotics. J. Mol. Biol. 359, 728–740 (2006)
Frerichs-Deeken, U. et al. Dioxygenases without requirement for cofactors and their chemical model reaction: compulsory order ternary complex mechanism of 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase involving general base catalysis by histidine 251 and single-electron oxidation of the substrate dianion. Biochemistry 43, 14485–14499 (2004)
Massey, V. Activation of molecular oxygen by flavins and flavoproteins. J. Biol. Chem. 269, 22459–22462 (1994)
Roth, J. P. & Klinman, J. P. Catalysis of electron transfer during activation of O2 by the flavoprotein glucose oxidase. Proc. Natl Acad. Sci. USA 100, 62–67 (2003)
Pootoolal, J. et al. Assembling the glycopeptide antibiotic scaffold: The biosynthesis of A47934 from Streptomyces toyocaensis NRRL15009. Proc. Natl Acad. Sci. USA 99, 8962–8967 (2002)
Otwinowski, Z. & Minor, W. in Methods in Enzymology: Macromolecular Crystallography Vol. 276 (eds Sweet, R. M. & Carter, C. W.) Part A 307–326 (Academic Press, New York, 1997)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, (Pt 12 Pt 1)2126–2132 (2004)
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Acknowledgements
We are grateful to A. Heroux and the staff at the Brookhaven National Synchrotron Light Source PXRR for assistance with X-ray data collection and processing. We are grateful to C. C. Tseng, R. S. Klausen and C. T. Walsh for discussions and experimental assistance. We thank P. D. Fortin and A. R. Howard-Jones for assistance in oxygen-binding measurements, Z. Dauter for advice on X-ray data analysis, and members of the Bruner research group and Boston College Chemistry Department for helpful discussions and comments on the manuscript. This work was supported with funds from Boston College and the Damon Runyon Cancer Research Foundation.
Author Contributions P.F.W. crystallized and solved the structure of apo-DpgC. E.N.F. crystallized the DpgC/inhibitor co-complex and with P.F.W. solved the structure. E.N.F constructed and with P.F.W. performed biochemical assays on mutant DpgC enzymes. Y.L. synthesized and biochemically evaluated the inhibitor, DPA-NH-CoA. S.D.B. wrote the manuscript and all authors discussed the results and commented on the manuscript.
The atomic coordinates for the DpgC/inhibitor complex structure have been deposited in the Protein Data Bank with accession code 2NP9.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures S1–S4, Supplementary Table 1 and Supplementary Methods. The Supplementary Figures illustrate additional electron density maps, plots of kinetic data characterizing the synthetic inhibitor and mutant enzymes, experimental data for determination of multimeric state and a plot of data from O2 kinetic binding experiments. A Supplementary Table is included with crystallographic statistics. The Supplementary Methods section describes the cloning and purification of DpgC, enzyme kinetic assays, site-directed mutagenesis, molecular weight determination and synthesis of the inhibitor DPA-NH-CoA. (PDF 1123 kb)
Rights and permissions
About this article
Cite this article
Widboom, P., Fielding, E., Liu, Y. et al. Structural basis for cofactor-independent dioxygenation in vancomycin biosynthesis. Nature 447, 342–345 (2007). https://doi.org/10.1038/nature05702
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature05702
This article is cited by
-
RETRACTED ARTICLE: Endoperoxide formation by an α-ketoglutarate-dependent mononuclear non-haem iron enzyme
Nature (2015)
-
Stereoselective C–C bond formation catalysed by engineered carboxymethylproline synthases
Nature Chemistry (2011)
-
Cofactor-independent oxidases and oxygenases
Applied Microbiology and Biotechnology (2010)
-
Cofactor-independent oxygenases go it alone
Nature Chemical Biology (2007)
-
Research highlights
Nature (2007)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.