Abstract
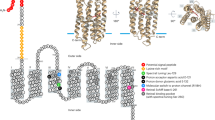
Proteorhodopsins are bacterial light-dependent proton pumps. Their discovery within genomic material from uncultivated marine bacterioplankton caused considerable excitement because it indicated a potential phototrophic function within these organisms, which had previously been considered strictly chemotrophic1. Subsequent studies established that sequences encoding proteorhodopsin are broadly distributed throughout the world’s oceans2,3,4,5. Nevertheless, the role of proteorhodopsins in native marine bacteria is still unknown6. Here we show, from an analysis of the complete genomes of three marine Flavobacteria, that cultivated bacteria in the phylum Bacteroidetes, one of the principal components of marine bacterioplankton, contain proteorhodopsin. Moreover, growth experiments in both natural and artificial seawater (low in labile organic matter, which is typical of the world’s oceans) establish that exposure to light results in a marked increase in the cell yield of one such bacterium (Dokdonia sp. strain MED134) when compared with cells grown in darkness. Thus, our results show that the phototrophy conferred by proteorhodopsin can provide critical amounts of energy, not only for respiration and maintenance but also for active growth of marine bacterioplankton in their natural environment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Béjà, O. et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289, 1902–1906 (2000)
Béjà, O., Spudich, E. N., Spudich, J. L., Leclerc, M. & DeLong, E. F. Proteorhodopsin phototrophy in the ocean. Nature 411, 786–789 (2001)
de la Torre, J. R. et al. Proteorhodopsin genes are distributed among divergent marine bacterial taxa. Proc. Natl Acad. Sci. USA 100, 12830–12835 (2003)
Sabehi, G. et al. Novel proteorhodopsin variants from the Mediterranean and Red Seas. Environ. Microbiol. 5, 842–849 (2003)
Venter, J. C. et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74 (2004)
Giovannoni, S. J. et al. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature 438, 82–85 (2005)
Man, D. et al. Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 22, 1725–1731 (2003)
Sabehi, G. et al. New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol. 3, e273 (2005)
Giovannoni, S. & Rappé, M. in Microbial Ecology of the Oceans (ed. Kirchman, D.) 47–84 (Wiley-Liss, New York, 2000)
Glöckner, F. O., Fuchs, B. M. & Amann, R. Bacterioplankton composition of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65, 3721–3726 (1999)
Hagström, Å. et al. Use of 16S ribosomal DNA for delineation of marine bacterioplankton species. Appl. Environ. Microbiol. 68, 3628–3633 (2002)
Kirchman, D. L. The ecology of Cytophaga–Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39, 91–100 (2002)
Rappé, M. S., Connon, S. A., Vergin, K. L. & Giovannoni, S. J. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418, 630–633 (2002)
Dioumaev, A. K. et al. Proton transfers in the photochemical reaction cycle of proteorhodopsin. Biochemistry 41, 5348–5358 (2002)
Wang, W.-W., Sineshchekov, O. A., Spudich, E. N. & Spudich, J. L. Spectroscopic and photochemical characterization of a deep ocean proteorhodopsin. J. Biol. Chem. 278, 33985–33991 (2003)
Neutze, R. et al. Bacteriorhodopsin: a high-resolution structural view of vectorial proton transport. Biochim. Biophys. Acta 1565, 144–167 (2002)
Teramoto, M., Takaichi, S., Inomata, Y., Ikenaga, H. & Misawa, N. Structural and functional analysis of a lycopene L-monocyclase gene isolated from a unique marine bacterium that produces myxol. FEBS Lett. 545, 120–126 (2003)
Abell, G. C. J. & Bowman, J. P. Ecological and biogeographic relationships of class Flavobacteria in the Southern Ocean. FEMS Microbiol. Ecol. 51, 265–277 (2005)
Pinhassi, J. et al. Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70, 6753–6766 (2004)
Riemann, L., Steward, G. F. & Azam, F. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66, 578–587 (2000)
Shiba, T., Simidu, U. & Taga, N. Distribution of aerobic bacteria which contain bacteriochlorophyll a. Appl. Environ. Microbiol. 38, 43–48 (1979)
Kolber, Z. S., Van Dover, C. L., Niederman, R. A. & Falkowski, P. G. Bacterial photosynthesis in surface waters of the open ocean. Nature 407, 177–179 (2000)
Béjà, O. et al. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415, 630–633 (2002)
Bielawski, J. P., Dunn, K. A., Sabehi, G. & Béjà, O. Darwinian adaptation of proteorhodopsin to different light intensities in the marine environment. Proc. Natl Acad. Sci. USA 101, 14824–14829 (2004)
Frigaard, N.-U., Martinez, A., Mincer, T. J. & DeLong, E. F. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature 439, 847–850 (2006)
Sabehi, G., Béjà, O., Suzuki, M. T., Preston, C. M. & DeLong, E. F. Different SAR86 subgroups harbour divergent proteorhodopsins. Environ. Microbiol. 6, 903–910 (2004)
Acknowledgements
We thank S. Arnautovic, J. O. Ekström, M. Widell, E. Lundberg and E. Lindehoff for help with growth experiments, ultracentrifugation, cloning, dissolved organic carbon and nutrient analysis, respectively, and T. Berman for helpful comments on the manuscript. We thank the Swedish Science Council, the Spanish Ministerio de Educación y Ciencia, Swegene, EMEP, and SSF for supporting this research.
The genomes of strains MED134, MED152 and MED217 are deposited in GenBank under accession numbers AAMZ00000000, AANA00000000 and AANC00000000, and their 16S rRNA gene sequences under accession numbers DQ481462, DQ481463 and DQ294290, respectively. The amino acid sequences of MED134 and MED152 PR are deposited in GenBank under accession numbers ZP_01049273 and ZP_01054176.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains the following: Supplementary Figures and Legends 1-3; Supplementary Methods; Supplementary Notes. The Supplementary Figures show proteorhodopsin amino acid sequence alignment, laser-flash induced absorbance changes of proteorhodopsin in membrane preparation of Dokdonia sp. MED134 and arrangement of proteorhodopsin, ß-carotene and retinal synthesis genes. (PDF 1576 kb)
Rights and permissions
About this article
Cite this article
Gómez-Consarnau, L., González, J., Coll-Lladó, M. et al. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature 445, 210–213 (2007). https://doi.org/10.1038/nature05381
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature05381
This article is cited by
-
A comparative study reveals the relative importance of prokaryotic and eukaryotic proton pump rhodopsins in a subtropical marginal sea
ISME Communications (2023)
-
Rhodopsin-mediated nutrient uptake by cultivated photoheterotrophic Verrucomicrobiota
The ISME Journal (2023)
-
Microbial degradation of various types of dissolved organic matter in aquatic ecosystems and its influencing factors
Science China Earth Sciences (2023)
-
Metagenomic analysis reveals wide distribution of phototrophic bacteria in hydrothermal vents on the ultraslow-spreading Southwest Indian Ridge
Marine Life Science & Technology (2022)
-
Metapangenomics reveals depth-dependent shifts in metabolic potential for the ubiquitous marine bacterial SAR324 lineage
Microbiome (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.