Abstract
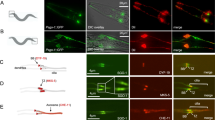
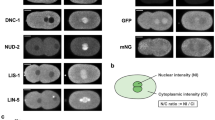
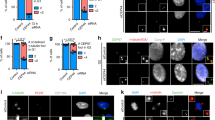
Centrioles are necessary for flagella and cilia formation1,2, cytokinesis3,4, cell-cycle control5 and centrosome organization/spindle assembly6. They duplicate once per cell cycle, but the mechanisms underlying their duplication remain unclear. Here we show using electron tomography of staged C. elegans one-cell embryos that daughter centriole assembly begins with the formation and elongation of a central tube followed by the peripheral assembly of nine singlet microtubules. Tube formation and elongation is dependent on the SAS-5 and SAS-6 proteins, whereas the assembly of singlet microtubules onto the central tube depends on SAS-4. We further show that centriole assembly is triggered by an upstream signal mediated by SPD-2 and ZYG-1. These results define a structural pathway for the assembly of a daughter centriole and should have general relevance for future studies on centriole assembly in other organisms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dutcher, S. K. Elucidation of basal body and centriole functions in Chlamydomonas reinhardtii. Traffic 4, 443–451 (2003)
Briggs, L. J., Davidge, J. A., Wickstead, B., Ginger, M. L. & Gull, K. More than one way to build a flagellum: comparative genomics of parasitic protozoa. Curr. Biol. 14, R611–R612 (2004)
Gromley, A. et al. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell 123, 75–87 (2005)
Piel, M., Nordberg, J., Euteneuer, U. & Bornens, M. Centrosome-dependent exit of cytokinesis in animal cells. Science 291, 1550–1553 (2001)
Doxsey, S., McCollum, D. & Theurkauf, W. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 21, 411–434 (2005)
Sluder, G. & Rieder, C. L. Centriole number and the reproductive capacity of spindle poles. J. Cell Biol. 100, 887–896 (1985)
Azimzadeh, J. & Bornens, M. in Centrosomes in Development and Disease (ed. Nigg, E. A.) 93–122 (Wiley-VCH, Weinheim, 2004)
Dippell, R. V. The development of basal bodies in paramecium. Proc. Natl Acad. Sci. USA 61, 461–468 (1968)
Vorobjev, I. A. & Chentsov Yu, S. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 93, 938–949 (1982)
O'Toole, E. T. et al. Morphologically distinct microtubule ends in the mitotic centrosome of Caenorhabditis elegans. J. Cell Biol. 163, 451–456 (2003)
Gönczy, P. et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408, 331–336 (2000)
Sönnichsen, B. et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434, 462–469 (2005)
Zipperlen, P., Fraser, A. G., Kamath, R. S., Martinez-Campos, M. & Ahringer, J. Roles for 147 embryonic lethal genes on C. elegans chromosome I identified by RNA interference and video microscopy. EMBO J. 20, 3984–3992 (2001)
O'Connell, K. F. et al. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell 105, 547–558 (2001)
O'Connell, K. F., Leys, C. M. & White, J. G. A genetic screen for temperature-sensitive cell-division mutants of Caenorhabditis elegans. Genetics 149, 1303–1321 (1998)
O'Connell, K. F., Maxwell, K. N. & White, J. G. The spd-2 gene is required for polarization of the anteroposterior axis and formation of the sperm asters in the Caenorhabditis elegans zygote. Dev. Biol. 222, 55–70 (2000)
Pelletier, L. et al. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 14, 863–873 (2004)
Leidel, S. & Gönczy, P. SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell 4, 431–439 (2003)
Leidel, S. & Gönczy, P. Centrosome duplication and nematodes: recent insights from an old relationship. Dev. Cell 9, 317–325 (2005)
Kirkham, M., Müller-Reichert, T., Oegema, K., Grill, S. & Hyman, A. A. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112, 575–587 (2003)
Kemp, C. A., Kopish, K. R., Zipperlen, P., Ahringer, J. & O'Connell, K. F. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell 6, 511–523 (2004)
Delattre, M. et al. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nature Cell Biol. 6, 656–664 (2004)
Dammermann, A. et al. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell 7, 815–829 (2004)
Leidel, S., Delattre, M., Cerutti, L., Baumer, K. & Gönczy, P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nature Cell Biol. 7, 115–125 (2005)
Müller-Reichert, T., Srayko, M., Hyman, A., O'Toole, E. & McDonald, K. Correlative light and electron microscopy of early Caenorhabditis elegans embryos in mitosis. Meth. Cell Biol. 79, 99–117 (in the press)
Kuriyama, R. & Borisy, G. G. Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. J. Cell Biol. 91, 814–821 (1981)
Robbins, E., Jentzsch, G. & Micali, A. The centriole cycle in synchronized HeLa cells. J. Cell Biol. 36, 329–339 (1968)
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996)
Delattre, M., Canard, C. & Gönczy, P. Sequential protein recruitment of C. elegans centriole formation. Curr. Biol. 16, 1844–1849 (2006)
Acknowledgements
We thank A. Dammermann and K. Oegema for their gift of the GFP–ZYG-1 strain and for sharing unpublished results; F. Barr for anti-GFP antibodies; M. Tipsword, M. Ruer and D. Drechsel for generating the ZYG-1 antibodies; A. Pozniakovsky for generating the GFP–SAS-5 and GFP–SAS-6 constructs; O. Molodtsova for biolistic bombardment; J. Mäntler and Q. DeRobillard for help with electron microscopy; C. Lorenz for secretarial assistance; K. McDonald, P. Verkade and I. Baines for help in developing the single-embryo high-pressure freezing approach; C. Eckmann for providing the FOG-1 feeding construct; and F. Friedrich for help with graphical artwork. We are grateful to G. Warren, W. Zacheriae, R. Kittler and F. Quittnat Pelletier for their comments on the manuscript, and Hyman laboratory members for stimulating discussions. The Boulder Laboratory for 3D Electron Microscopy of Cells is supported by a grant from the National Institutes of Health to J. R. McIntosh. L.P. is supported by a postdoctoral fellowship from the HFSP.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Methods
This file contains additional details of the methods used in this study. (DOC 152 kb)
Supplementary Movie
This file contains text to accompany the below Supplementary Movies. (DOC 30 kb)
Supplementary Figure Legends
This file contains text to accompany the below Supplementary Figures. (DOC 30 kb)
Supplementary Figure 1
In glutharaldehyde-fixed C. elegans embryos daughter centrioles are first observed at pronuclear migration. (JPG 1431 kb)
Supplementary Figure 2
Mating assays can be used to monitor the recruitment of centriole proteins to the site of daughter centriole assembly. (JPG 58 kb)
Supplementary Figure 3
ZYG-1::GFP is recruited during meiosis. (PDF 8162 kb)
Supplementary Figure s4
Electron tomography of staged one-cell embryos. (JPG 40 kb)
Supplementary Figure s5
Quantification of 3D centriole models (PDF 3133 kb)
Supplementary Figure s6
The use of the slicer tool to display centriole structure. (JPG 195 kb)
Supplementary Figure s7
Modelling centriole structures in 3dmod. (JPG 150 kb)
Movie Figure 3A1.
Complete tomographic volume and three-dimensional model of centrioles at pronuclear appearance. (MOV 12527 kb)
Movie Figure 3A2.
Complete tomographic volume and three-dimensional model of centrioles at pronuclear appearance. (MOV 5058 kb)
Movie Figure 3B1
Complete tomographic volume and three-dimensional model of centrioles during pronuclear migration. (MOV 5982 kb)
Movie Figure 3B2
Complete tomographic volume and three-dimensional model of centrioles during pronuclear migration. (MOV 8137 kb)
Movie Figure 3C1
Complete tomographic volume and three-dimensional model of centrioles during the rotation of the pronuclei/centrosome complex. (MOV 3718 kb)
Movie Figure 3C2
Complete tomographic volume and three-dimensional model of centrioles during the rotation of the pronuclei/centrosome complex. (MOV 6824 kb)
Movie Figure 4A
Complete tomographic volume and three-dimensional model of a zyg-1(RNAi) embryo during the rotation of the pronuclei/centrosome complex. (MOV 6467 kb)
Movie Figure 4B
Complete tomographic volume and three-dimensional model of a sas-6(RNAi) embryo during rotation of the pronuclei/centrosome complex (MOV 3422 kb)
Movie Figure 4C
Complete tomographic volume and three-dimensional model of a sas-4(RNAi) embryo at pronuclear appearance. (MOV 8180 kb)
Movie Slicer Tool 1
Movie through serial tomographic slices showing a mother centriole in cross section and a daughter central tube during pronuclear migration. (MOV 6256 kb)
Movie Slicer Tool 2
Movie through serial tomographic slices of the data set in “Movie Slicer Tool 1” rotated to display the mother centriole along the longitudinal axis. (MOV 4011 kb)
Movie SAS-5(RNAi).
Complete tomographic volume and three-dimensional model of a sas-5(RNAi) embryo during rotation of the pronuclei/centrosome complex. (MOV 14434 kb)
Movie SAS-4(RNAi).
Complete tomographic volume and three-dimensional model of a sas-4(RNAi) embryo during rotation of the pronuclei/centrosome complex. (MOV 23365 kb)
Supplementary Table 1
Quantitative analysis of centriole assembly in one-cell wild-type and RNAi embryos. (DOC 88 kb)
Rights and permissions
About this article
Cite this article
Pelletier, L., O’Toole, E., Schwager, A. et al. Centriole assembly in Caenorhabditis elegans. Nature 444, 619–623 (2006). https://doi.org/10.1038/nature05318
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05318
This article is cited by
-
ELI trifocal microscope: a precise system to prepare target cryo-lamellae for in situ cryo-ET study
Nature Methods (2023)
-
Centrosome: A Microtubule Nucleating Cellular Machinery
Journal of the Indian Institute of Science (2021)
-
STIL: a multi-function protein required for dopaminergic neural proliferation, protection, and regeneration
Cell Death Discovery (2019)
-
Direct binding of CEP85 to STIL ensures robust PLK4 activation and efficient centriole assembly
Nature Communications (2018)
-
The rise and fall of basal bodies in the nematode Caenorhabditis elegans
Cilia (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.