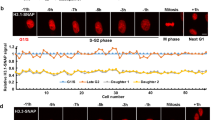
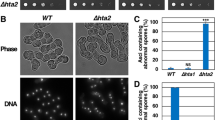
Abstract
Proper histone levels are critical for transcription, chromosome segregation, and other chromatin-mediated processes1–7. In Saccharomyces cerevisiae, the histones H2A and H2B are encoded by two gene pairs, named HTA1-HTB1 and HTA2-HTB2 (ref. 8). Previous studies have demonstrated that when HTA2-HTB2 is deleted, HTA1-HTB1 dosage compensates at the transcriptional level4,9. Here we show that a different mechanism of dosage compensation, at the level of gene copy number, can occur when HTA1-HTB1 is deleted. In this case, HTA2-HTB2 amplifies via creation of a new, small, circular chromosome. This duplication, which contains 39 kb of chromosome II, includes HTA2-HTB2, the histone H3-H4 locus HHT1-HHF1, a centromere and origins of replication. Formation of the new chromosome occurs by recombination between two Ty1 retrotransposon elements that flank this region. Following meiosis, recombination between these two particular Ty1 elements occurs at a greatly elevated level in hta1-htb1Δ mutants, suggesting that a decreased level of histones H2A and H2B specifically stimulates this amplification of histone genes. Our results demonstrate another mechanism by which histone gene dosage is controlled to maintain genomic integrity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Meeks-Wagner, D. & Hartwell, L. H. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 44, 43–52 (1986)
Gunjan, A. & Verreault, A. A. Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae.. Cell 115, 537–549 (2003)
Norris, D., Dunn, B. & Osley, M. A. The effect of histone gene deletions on chromatin structure in Saccharomyces cerevisiae.. Science 242, 759–761 (1988)
Norris, D. & Osley, M. A. The two gene pairs encoding H2A and H2B play different roles in the Saccharomyces cerevisiae life cycle. Mol. Cell. Biol. 7, 3473–3481 (1987)
Han, M., Chang, M., Kim, U. J. & Grunstein, M. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell 48, 589–597 (1987)
Clark-Adams, C. D., Norris, D., Osley, M. A., Fassler, J. S. & Winston, F. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 2, 150–159 (1988)
Han, M. & Grunstein, M. Nucleosome loss activates yeast downstream promoters in vivo.. Cell 55, 1137–1145 (1988)
Hereford, L., Fahrner, K., Woolford, J., Rosbash, M. & Kaback, D. B. Isolation of yeast histone genes H2A and H2B. Cell 18, 1261–1271 (1979)
Moran, L., Norris, D. & Osley, M. A. A yeast H2A–H2B promoter can be regulated by changes in histone gene copy number. Genes Dev. 4, 752–763 (1990)
Rykowski, M. C., Wallis, J. W., Choe, J. & Grunstein, M. Histone H2B subtypes are dispensable during the yeast cell cycle. Cell 25, 477–487 (1981)
Kolodrubetz, D., Rykowski, M. C. & Grunstein, M. Histone H2A subtypes associate interchangeably in vivo with histone H2B subtypes. Proc. Natl Acad. Sci. USA 79, 7814–7818 (1982)
Hirschhorn, J. N., Brown, S. A., Clark, C. D. & Winston, F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 6, 2288–2298 (1992)
Hirschhorn, J. N., Bortvin, A. L., Ricupero-Hovasse, S. L. & Winston, F. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo.. Mol. Cell. Biol. 15, 1999–2009 (1995)
Giaever, G. et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 (2002)
Struhl, K., Stinchcomb, D. T., Scherer, S. & Davis, R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc. Natl Acad. Sci. USA 76, 1035–1039 (1979)
Kupiec, M. & Petes, T. D. Allelic and ectopic recombination between Ty elements in yeast. Genetics 119, 549–559 (1988)
Kupiec, M. & Petes, T. D. Meiotic recombination between repeated transposable elements in Saccharomyces cerevisiae.. Mol. Cell. Biol. 8, 2942–2954 (1988)
Roeder, G. S. Unequal crossing-over between yeast transposable elements. Mol. Gen. Genet. 190, 117–121 (1983)
Ben-Aroya, S., Mieczkowski, P. A., Petes, T. D. & Kupiec, M. The compact chromatin structure of a Ty repeated sequence suppresses recombination hotspot activity in Saccharomyces cerevisiae.. Mol. Cell 15, 221–231 (2004)
Petes, T. D. Meiotic recombination hot spots and cold spots. Nature Rev. Genet. 2, 360–369 (2001)
Wu, X. & Haber, J. E. A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell 87, 277–285 (1996)
Ercan, S., Reese, J. C., Workman, J. L. & Simpson, R. T. Yeast recombination enhancer is stimulated by transcription activation. Mol. Cell. Biol. 25, 7976–7987 (2005)
Zeyl, C. Capturing the adaptive mutation in yeast. Res. Microbiol. 155, 217–223 (2004)
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001)
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001)
Schwartz, D. C. & Cantor, C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 37, 67–75 (1984)
Acknowledgements
We thank A. Dudley and D. Helmlinger for helpful comments on the manuscript. We also thank J. Haber and J.-A. Kim for the suggestion of and advice on the HO experiment, and V. Dror for instruction about CHEF gels. We are grateful to A. Gabriel, P. Kaufman, M. A. Osley and T. Petes for sharing unpublished results and for discussions. We thank J. Hirschhorn, whose observations led to this project. This work was supported by a grant from the National Institutes of Health to F.W. and by a National Science Foundation Graduate Fellowship to D.E.L.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
This file contains Supplementary Discussion, Supplementary Methods, Supplementary Tables 1–6 and Supplementary Figures 1–7. (PDF 6095 kb)
Rights and permissions
About this article
Cite this article
Libuda, D., Winston, F. Amplification of histone genes by circular chromosome formation in Saccharomyces cerevisiae. Nature 443, 1003–1007 (2006). https://doi.org/10.1038/nature05205
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature05205
This article is cited by
-
Revisiting characteristics of oncogenic extrachromosomal DNA as mobile enhancers on neuroblastoma and glioma cancers
Cancer Cell International (2022)
-
Small ring has big potential: insights into extrachromosomal DNA in cancer
Cancer Cell International (2021)
-
Extrachromosomal circular DNA: a new potential role in cancer progression
Journal of Translational Medicine (2021)
-
Phylogeny, evolution, and potential ecological relationship of cytochrome CYP52 enzymes in Saccharomycetales yeasts
Scientific Reports (2020)
-
Circular DNA elements of chromosomal origin are common in healthy human somatic tissue
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.