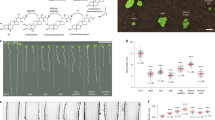
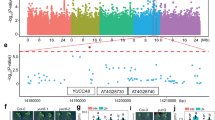
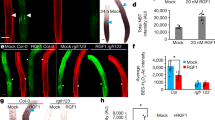
Abstract
Brassinosteroid and auxin decisively influence plant development, and overlapping transcriptional responses to these phytohormones suggest an interaction between the two pathways1,2,3. However, whether this reflects direct feedback or merely parallel inputs on common targets is unclear. Here we show that in Arabidopsis roots, this interaction is mediated by BREVIS RADIX (BRX), which is required for optimal root growth4. We demonstrate that the brx phenotype results from a root-specific deficiency of brassinosteroid and is due to reduced, BRX-dependent expression of a rate-limiting enzyme in brassinosteroid biosynthesis. Unexpectedly, this deficiency affects the root expression level of ∼15% of all Arabidopsis genes, but the transcriptome profile can be restored to wild type by brassinosteroid treatment. Thus, proper brassinosteroid levels are required for the correct expression of many more genes than previously suspected. Moreover, embryonic or post-embryonic brassinosteroid application fully or partially, respectively, rescues the brx phenotype. Further, auxin-responsive gene expression is globally impaired in brx, demonstrating that brassinosteroid levels are rate-limiting for auxin-responsive transcription. BRX expression is strongly induced by auxin and mildly repressed by brassinolide, which means that BRX acts at the nexus of a feedback loop that maintains threshold brassinosteroid levels to permit optimal auxin action.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Goda, H. et al. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 134, 1555–1573 (2004)
Nemhauser, J. L., Mockler, T. C. & Chory, J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2, E258 (2004)
Zurek, D. M., Rayle, D. L., McMorris, T. C. & Clouse, S. D. Investigation of gene expression, growth kinetics, and wall extensibility during brassinosteroid-regulated stem elongation. Plant Physiol. 104, 505–513 (1994)
Mouchel, C. F., Briggs, G. C. & Hardtke, C. S. Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev. 18, 700–714 (2004)
Briggs, G. C., Mouchel, C. F. & Hardtke, C. S. Characterization of the plant-specific BREVIS RADIX gene family reveals limited genetic redundancy despite high sequence conservation. Plant Physiol. 140, 1306–1316 (2006)
Blilou, I. et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44 (2005)
Friml, J., Wiśniewska, J., Benková, E., Mendgen, K. & Palme, K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809 (2002)
Terasaka, K. et al. PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell 17, 2922–2939 (2005)
Paciorek, T. et al. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435, 1251–1256 (2005)
Szekeres, M. et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182 (1996)
Tanaka, K. et al. Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol. 138, 1117–1125 (2005)
Mussig, C., Fischer, S. & Altmann, T. Brassinosteroid-regulated gene expression. Plant Physiol. 129, 1241–1251 (2002)
Shimada, Y. et al. Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol. 131, 287–297 (2003)
Bancos, S. et al. Regulation of transcript levels of the Arabidopsis cytochrome P450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 130, 504–513 (2002)
Beemster, G. T. & Baskin, T. I. Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 116, 1515–1526 (1998)
Beemster, G. T. & Baskin, T. I. STUNTED PLANT 1 mediates effects of cytokinin, but not of auxin, on cell division and expansion in the root of Arabidopsis. Plant Physiol. 124, 1718–1727 (2000)
Goda, H., Shimada, Y., Asami, T., Fujioka, S. & Yoshida, S. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 130, 1319–1334 (2002)
Vert, G., Nemhauser, J. L., Geldner, N., Hong, F. & Chory, J. Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 21, 177–201 (2005)
Wang, S., Tiwari, S. B., Hagen, G. & Guilfoyle, T. J. AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 17, 1979–1993 (2005)
Nakamura, A. et al. Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol. 133, 1843–1853 (2003)
Nakamura, A. et al. Arabidopsis Aux/IAA genes are involved in brassinosteroid-mediated growth responses in a manner dependent on organ type. Plant J. 45, 193–205 (2006)
Leyser, H. M. Molecular genetics of auxin signaling. Annu. Rev. Plant Biol. 53, 377–398 (2002)
Friml, J. et al. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108, 661–673 (2002)
Sabatini, S. et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99, 463–472 (1999)
Sieberer, T., Hauser, M. T., Seifert, G. J. & Luschnig, C. PROPORZ1, a putative Arabidopsis transcriptional adaptor protein, mediates auxin and cytokinin signals in the control of cell proliferation. Curr. Biol. 13, 837–842 (2003)
Tsugeki, R. & Fedoroff, N. V. Genetic ablation of root cap cells in Arabidopsis. Proc. Natl Acad. Sci. USA 96, 12941–12946 (1999)
Birnbaum, K. et al. A gene expression map of the Arabidopsis root. Science 302, 1956–1960 (2003)
Himanen, K. et al. Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14, 2339–2351 (2002)
Irizarry, R. A. et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 (2003)
Boyes, D. C. et al. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13, 1499–1510 (2001)
Acknowledgements
We thank T. Guilfoyle for the DR5::GUS reporter line; the DNA Array Facility of the University of Lausanne for assistance with microarray experiments; and J. MacDonald Chambers-Petetôt for assistance with sectioning and microscopy. C.F.M. was supported by the Canton de Vaud, Switzerland. This work was funded by a grant from the Swiss National Science Foundation awarded to C.S.H. Author Contributions C.F.M. contributed all data except those for Fig. 2c and 2h, and Supplementary Figs 1i and 3, contributed by K.S.O. All authors were involved in the design of the research and the writing of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The microarray experiments have been deposited in the Array Express Database under accession numbers E-MEXP-635 and M-MEXP-637. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Figure 1
Expression and requirement of BRX in the root vasculature. (JPG 3232 kb)
Supplementary Figure 2
Expression level ratios for various brassinosteroid biosynthesis or signaling genes extracted from the various microarray experiments. (JPG 559 kb)
Supplementary Figure 3
Root growth in bri1 mutants. (JPG 482 kb)
Supplementary Figure 4
Predicted transcription factor binding sites in the BRX promoter (JPG 237 kb)
Supplementary Figure 5
Instability of BRX-GFP fusion protein. (JPG 472 kb)
Supplementary Table
Expression ratios of differentially expressed genes in microarray analyses. Original microarray data have been deposited at ArrayExpress database. (XLS 2748 kb)
Rights and permissions
About this article
Cite this article
Mouchel, C., Osmont, K. & Hardtke, C. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443, 458–461 (2006). https://doi.org/10.1038/nature05130
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature05130
This article is cited by
-
Transcriptome analysis reveals the effect of acidic environment on adventitious root differentiation in Camellia sinensis
Plant Molecular Biology (2023)
-
Dehydration stress influences the expression of brevis radix gene family members in sorghum (Sorghum bicolor)
Proceedings of the Indian National Science Academy (2022)
-
Transcriptome analysis revealed the regulation of gibberellin and the establishment of photosynthetic system promote rapid seed germination and early growth of seedling in pearl millet
Biotechnology for Biofuels (2021)
-
Genome-wide expression and network analyses of mutants in key brassinosteroid signaling genes
BMC Genomics (2021)
-
Divergence of three BRX homoeologs in Brassica rapa and its effect on leaf morphology
Horticulture Research (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.